News archive
Foldit on the Web!
Introducing Foldit Education Mode v2 on the Web
In 2020, as the pandemic set in and classes moved online, the Foldit team realized they had a tremendous opportunity to help the science education community that was desperate for good science learning tools. They therefore created and released Foldit Education Mode in August 2020 as a ready-made teaching tool for biochemistry that instructors could use easily and freely in their courses. In the past five years, at least hundreds of educators and many thousands of students have used Foldit Education Mode.
While Foldit Education Mode was greatly successful at meeting the challenge of the moment, it still had flaws that would hold some students back from using it. The Foldit team also recently published a quantitative assessment of learning with Foldit Education Mode (https://doi.org/10.1002/bmb.21906), and while it was fulfilling its purpose, there was definitely also room to improve.
That’s why they're very excited to introduce their second release of Foldit Education Mode with improved levels and teaching tools, but even more excitingly: now available on the web! This means no more downloads required, and compatibility with any computer type. For many, this will enable use of Foldit in the classroom that was never previously possible, and for many more a great degree of increased flexibility and improved user experience.
To play Foldit Education Mode on the web, please go to https://play.fold.it/, and you can start playing immediately! You’ll notice several updates to the user interface along with the new and improved puzzles. The number of puzzles is slightly larger, but this change was largely made to break up levels that had too much biochemistry and gameplay content all rolled together, and by separating it out, the learning experience becomes considerably easier.
In the long term, the Foldit team plans to explore the possibility of hosting smaller Science puzzles to help bridge the gap for new players who start with the web version, but that will require more work to complete. In the meantime, they hope that educators and students worldwide will enjoy the new and improved experience, and those who couldn’t participate before will now be able to.
As always, the Foldit team would appreciate finding out who is using Foldit Education Mode, as they don’t collect that data; it really helps with procuring more funding to support these efforts. So if you do use it, please drop them a line at mail.fold.it@gmail.com.
This post originated from the Foldit blog post https://fold.it/forum/blog/introducing-foldit-education-mode-v2-on-the-web
11 Sep 2025, 22:35:26 UTC
· Discuss
Rosetta@home Update
First, we’d like to thank all our contributors who have supported and continue to contribute to our scientific projects through Rosetta@home. We want to update you on the status of Rosetta@home projects and our future plans.
With the advancement of AI models like AlphaFold and RosettaFold for protein structure predictions, Rosetta@home has been less used for this purpose. However, researchers are now utilizing Rosetta@home for small molecule and peptide designs, where even the current state-of-the-art AI models struggle due to limitations in generalizability to novel small molecules and non-canonical peptides.
Recently, we have developed a virtual screening protocol in Rosetta, named RosettaVS, for small molecule drug discovery. This work has been published in Nature Communications (https://doi.org/10.1038/s41467-024-52061-7), demonstrating that RosettaVS is one of the best physics-based virtual screening protocols. Combined with deep learning techniques, it can effectively screen multi-billion compound libraries and discover novel compounds for pharmaceutical targets.
While deep learning models like AlphaFold and RosettaFold can predict canonical peptide structures, they cannot handle peptides with non-canonical amino acids or mixed chirality. The physics-based force field in Rosetta has specialized terms to simulate these amino acids. Rosetta will be used to sample hundreds of thousands of different conformations of the designed peptide to validate the structure.
Looking ahead, Rosetta@home will be an invaluable platform for large-scale virtual screening and peptide simulations for drug discovery. We plan to launch more virtual screening jobs and peptide simulations on Rosetta@home in the near future.
Thank you!
4 Mar 2025, 4:50:22 UTC
· Discuss
COVID-19 vaccine with IPD nanoparticles wins full approval abroad
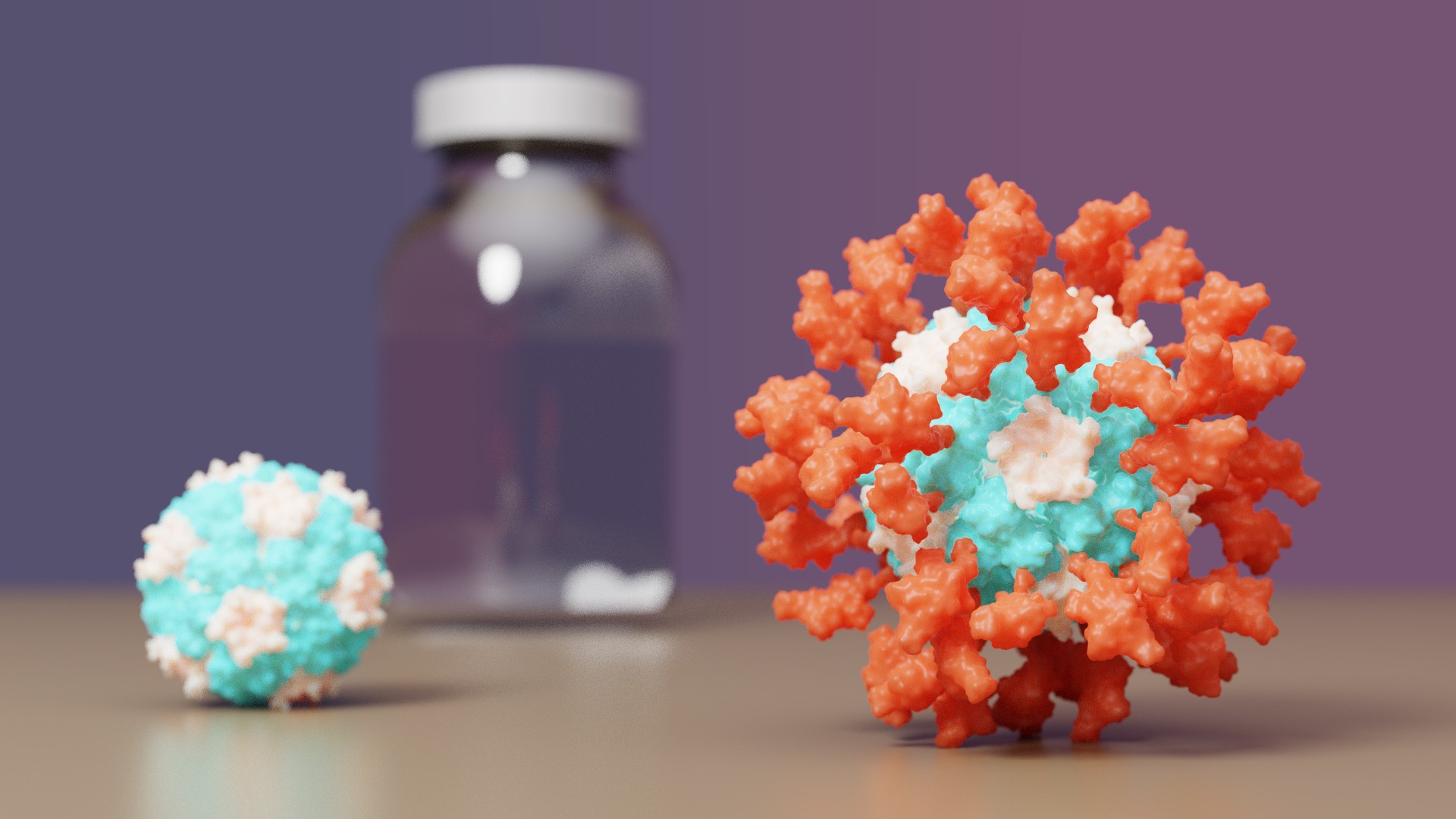
The IPD is excited to announce it's first designed protein medicine with full approval abroad.
Congrats and thank you to all Rosetta@home contributors! The computing you have provided has greatly aided in de novo protein design challenges such as vaccine development leading to breakthroughs like this.
For more information you can visit the IPD vaccine news post.
A video is also available here.
From the IPD news site:
• Clinical testing found the vaccine outperforms Oxford/AstraZeneca’s
• The protein-based vaccine, now called SKYCovione, does not require deep freezing
• University of Washington to waive royalty fees for the duration of the pandemic
• South Korea to purchase 10 million doses for domestic use
A vaccine for COVID-19 developed at the University of Washington School of Medicine has been approved by the Korean Ministry of Food and Drug Safety for use in individuals 18 years of age and older. The vaccine, now known as SKYCovione, was found to be more effective than the Oxford/AstraZeneca vaccine sold under the brand names Covishield and Vaxzevria.
SK bioscience, the company leading the SKYCovione’s clinical development abroad, is now seeking approval for its use in the United Kingdom and beyond. If approved by the World Health Organization, the vaccine will be made available through COVAX, an international effort to equitably distribute COVID-19 vaccines around the world. In addition, the South Korean government has agreed to purchase 10 million doses for domestic use.
The Seattle scientists behind the new vaccine sought to create a ‘second-generation’ COVID-19 vaccine that is safe, effective at low doses, simple to manufacture, and stable without deep freezing. These attributes could enable vaccination at a global scale by reaching people in areas where medical, transportation, and storage resources are limited.
“We know more than two billion people worldwide have not received a single dose of vaccine,” said David Veesler, associate professor of biochemistry at UW School of Medicine and co-developer of the vaccine. “If our vaccine is distributed through COVAX, it will allow it to reach people who need access.”
The University of Washington is licensing the vaccine technology royalty-free for the duration of the pandemic.
Congrats and thank you again to all R@h contributors!
6 Jul 2022, 18:27:59 UTC
· Discuss
Thank you!
We'd like to thank everyone who has contributed and continues to contribute to this project, and would like to remind everyone that there may be periods of down time while we are preparing for future large scale batches of jobs and analyzing results. We greatly appreciate your contributions which are vitally important to our ongoing research.
Thank you!
5 Nov 2020, 20:16:29 UTC
· Discuss
Coronavirus update from David Baker. Thank you all for your contributions!
Here is a short video of David Baker describing some exciting results from de novo designs targeting SARS-Cov-2.
Thank you all for your contributions to this research! Although R@h was not directly used for the work described in the publication (link provided below), R@h was used for designing relevant scaffolds. Additionally, there are currently many similar designs that bind SARS-Cov-2 and related targets that were engineered using R@h.
https://www.youtube.com/embed/ODEIN5V3yLg
More information is available from the publication, De novo design of picomolar SARS-CoV-2 mini protein inhibitors.
21 Sep 2020, 23:16:33 UTC
· Discuss
Switch to using SSL (Secure Socket Layer)
We updated our project to use SSL. The project URL has thus been changed to https://boinc.bakerlab.org/rosetta. You can reattach the project using this updated URL at your convenience. Please post any issues regarding this update in the discussion thread.
1 May 2020, 21:25:30 UTC
· Discuss
Help in the fight against COVID-19!
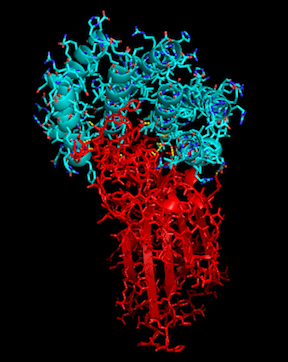
With the recent COVID-19 outbreak, R@h has been used to predict the structure of proteins important to the disease as well as to produce new, stable mini-proteins to be used as potential therapeutics and diagnostics, like the one displayed above which is bound to part of the SARS-CoV-2 spike protein.
To help our research, we are happy to announce a new application update, and thanks to the help from the Arm development community, including Rex St. John, Dmitry Moskalchuk, David Tischler, Lloyd Watts, and Sahaj Sarup, we are excited to also include the Linux-ARM platform. With this update we will continue to make protein binders to SARS-CoV-2 and related targets using the latest Rosetta source.
Thank you R@h volunteers for your continued support to this project. Your CPU hours are used not only to accurately model the structures of important proteins, but to design new ones as well. Let's band together and fight COVID-19!
More details will be available in the Discussion of this news post.
3 Apr 2020, 3:45:24 UTC
· Discuss
Designing shape-shifting proteins
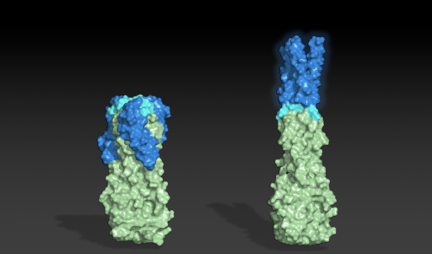
Thank you to all R@h participants who provided much of the computing used in a recent study published in PNAS describing the design of proteins that adopt more than one well-folded structure, reminiscent of viral fusion proteins.
For more infomation, click here.
19 Mar 2020, 0:47:01 UTC
· Discuss
Rosetta's role in fighting coronavirus
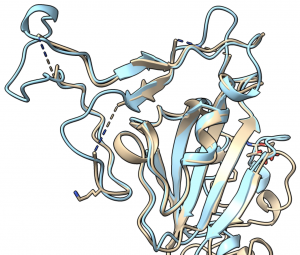
Thank you to all R@h volunteers for your contributions to help accurately model important coronavirus proteins. The collective computing power that you provide through R@h helps academic research groups world wide model important protein structures like these.
From a recent IPD news post:
"We are happy to report that the Rosetta molecular modeling suite was recently used to accurately predict the atomic-scale structure of an important coronavirus protein weeks before it could be measured in the lab. Knowledge gained from studying this viral protein is now being used to guide the design of novel vaccines and antiviral drugs."
Since the release of SARS-CoV-2 genome sequences in late January, a number of important corona virus proteins like the one described above have been modeled on R@h volunteer computers. A list of these proteins is provided by the Seattle Structural Genomics Center for Infectious Disease (SSGCID).
24 Feb 2020, 18:19:59 UTC
· Discuss
The Audacious Project

As you may have heard, the Institute for Protein Design was recently selected as part of The Audacious Project. This large-scale philanthropic collaboration, which is the successor to the TED Prize, surfaces and funds projects with the potential to change the world.
As a result, we are expanding our Seattle-based team of scientists and engineers who will work together to advance Rosetta, our software for protein design and structure prediction. The funding will also allow us to invest in the equipment, supplies and lab space needed to design and test millions of synthetic proteins.
What challenges will we be tackling? Watch my TED talk to find out.
All of this work — like everything we do — will depend on you, the participants in Rosetta@home. Whether it’s creating custom nanomaterials or safer cancer therapies, we rely on the Rosetta@home distributed computing platform. We cannot thank you enough for taking the time to be a part of this exciting research, and we hope you tell at least one friend that they too can play a role in the protein design revolution just by running Rosetta@home.
Thank you,
David Baker
Director, Institute for Protein Design
16 Jul 2019, 22:18:18 UTC
· Discuss
Coevolution at the proteome scale
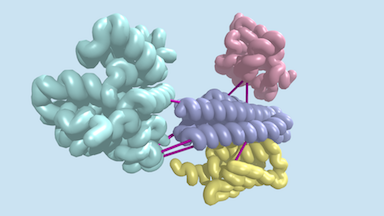
Last week, a report was published in Science describing the identification of hundreds of previously uncharacterized protein–protein interactions in E. coli and the pathogenic bacterium M. tuberculosis. These include both previously unknown protein complexes and previously uncharacterized components of known complexes. This research was led by postdoctoral fellow Qian Cong and included former Baker lab graduate student Sergey Ovchinnikov, now a John Harvard Distinguished Science Fellow at Harvard. Rosetta@home was used for much of the computing required for this work. Congratulations and thank you to all R@h volunteers.
For more information about this work click here.
15 Jul 2019, 19:23:58 UTC
· Discuss
Protein arrays on mineral surfaces

Last week, the Baker Lab in collaboration with the De Yoreo lab at PNNL published a report in Nature describing the design of synthetic protein arrays that assemble on the surface of mica, a common and exceptionally smooth crystalline mineral. This work provides a foundation for understanding how protein-crystal interactions can be systematically programmed. Although R@h was not directly used for this research, previously designed subunits were validated using R@h. Congratulations to all R@h volunteers and thank you for your continued contributions.
For more details click here.
15 Jul 2019, 19:12:28 UTC
· Discuss
Citizen scientists use Foldit to successfully design synthetic proteins
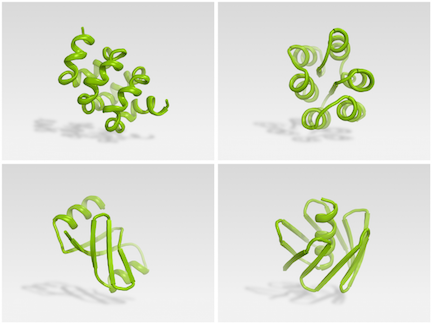

Citizen scientists can now use Foldit to successfully design synthetic proteins. The initial results of this unique collaboration are described in Nature.
Brian Koepnick, a recent PhD graduate in the Baker lab, led a team that worked on Foldit behind the scenes, introducing new features into the game that they believed would help players home in on better folded structures. Read more from the Baker Lab.
Thanks to all Rosetta@home participants who helped in this study. Many of the designs were validated using forward folding on Rosetta@home.
Read the full manuscript: https://doi.org/10.1038/s41586-019-1274-4 PDF
6 Jun 2019, 19:04:59 UTC
· Discuss
IPD's first nanoparticle vaccine
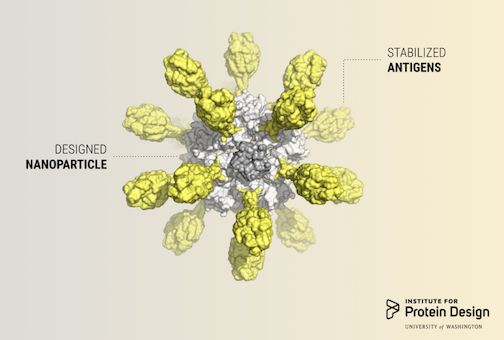
Researchers in the King lab, an affiliate of the Institute for Protein Design, published a report in Cell describing a computer-designed nanoparticle vaccine targeting respiratory syncytial virus (RSV). Although Rosetta@home was not directly used for this study, Rosetta@home volunteers provided computing for related research and development.
From IPD news:
Millions of children will visit hospitals this year, sickened by RSV. Infection is usually mild, causing only fevers, runny noses and frightened parents. But, in severe cases, barking coughs and painful wheezing can indicate serious respiratory complications, including bronchiolitis and pneumonia.
RSV is the primary cause of pneumonia in children under one and is therefore the leading cause of infant mortality worldwide after malaria. Although virtually every child on Earth will get RSV before the age of three, an estimated 99 percent of RSV deaths occur in developing countries. Despite substantial effort, there is not yet a safe and effective vaccine.
Today, an international team of scientists co-led by researchers at the IPD report in Cell a first-of-its-kind vaccine candidate for RSV. It elicits broadly neutralizing antibodies against respiratory syncytial virus in mice and monkeys, paving the way for human clinical trials.
8 Mar 2019, 18:51:56 UTC
· Discuss
Another publication in Nature describing the first de novo designed proteins with anti-cancer activity
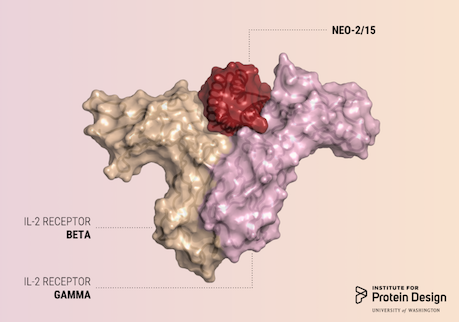
A report was published in Nature last week describing the first de novo designed proteins with anti-cancer activity.
These compact molecules were designed to stimulate the same receptors as IL-2, a powerful immunotherapeutic drug, while avoiding unwanted off-target receptor interactions. We believe this is just the first of many computer-generated cancer drugs with enhanced specificity and potency.
R@h participants provided computing for forward folding experiments used in this study which helped validate designs. We'd like to congratulate and thank all R@h volunteers who contributed to this work! Thank you!
Read the full article here: https://www.nature.com/articles/s41586-018-0830-7 (PDF)
14 Jan 2019, 22:58:59 UTC
· Discuss
Nature article on IPD work voted ‘2018 Reader’s Choice’
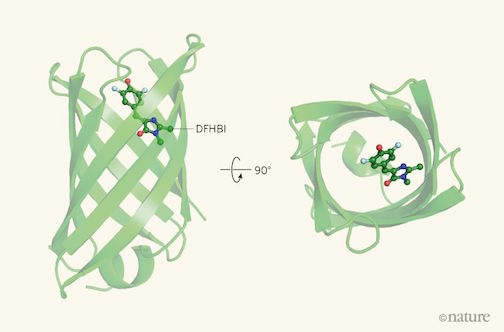
Readers of Nature’s News & Views selected an article about our work as their 2018 Reader’s Choice!
The article, written by Roberto Chica of University of Ottawa, does a fantastic job detailing our recent publication on de novo fluorescence-activating proteins — and the challenges of de novo protein design more generally.
4 Jan 2019, 1:06:51 UTC
· Discuss
New publication in Nature: programmable heterodimers
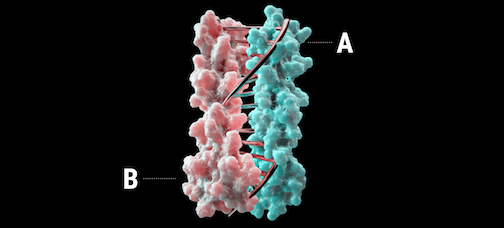
A new report was recently published in Nature describing the design of proteins that mimic DNA.
Using computational design, heterodimeric proteins that form double helices with hydrogen-bond mediated specificity were created. When a pool of these new protein zippers gets melted and then allowed to refold, only the proper pairings form. They are all-against-all orthogonal. With these new tools in hand, it may be possible to construct large protein-based machines that self-assemble in predictable ways.
Read the full article here: https://www.nature.com/articles/s41586-018-0802-y (PDF)
We'd like to thank all Rosetta@home volunteers who contributed computing resources used in this work. Thank you!
4 Jan 2019, 0:57:58 UTC
· Discuss
De novo design of self-assembling helical protein filaments
Another de novo design publication was released today describing the design of micron scale self-assembling helical filaments based on previously designed repeat proteins for which R@h participants contributed computing towards. Although R@h was not directly used for this study, R@h participants provided computing for related research. Thank you all for your continued contributions.
Read more here in Science.
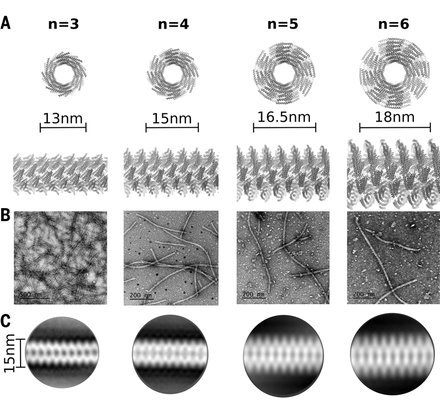
9 Nov 2018, 19:40:30 UTC
· Discuss
De novo design of jellyroll structures
Sorry for the late post. Last week the Baker lab and collaborators published the first example of proteins designed with non-local beta strand topology.
You can read more about this study here and the publication here.
Thanks to all Rosetta@home participants who contributed to this research.
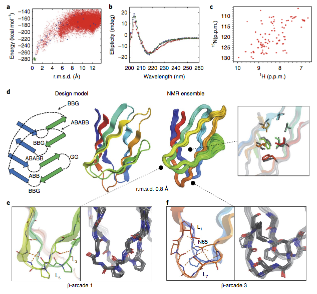
8 Nov 2018, 23:02:02 UTC
· Discuss
Discover magazine article about David Baker's progression into the field of protein design
An interesting article was released yesterday in Discover magazine about the historical progression of David Baker's research and how it has evolved into the Rosetta Commons community and the field of protein design. It includes a case example of de novo design research, and a perspective of what novel proteins may enable. Thank you all for your continued help and contributions to this research!
31 Oct 2018, 18:58:01 UTC
· Discuss
Rosetta 64 bit linux version 4.08 released
This update includes a fix suggested by rjs5 for the latest linux versions that use glibc 2.27+. Thank you rjs5!
Please post any issues/bugs regarding this app version in this thread.
2 Oct 2018, 17:46:01 UTC
· Discuss
Fluorescent proteins designed from scratch

Congrats to all Rosetta@home volunteers who contributed to a recent report in Nature describing the design of a completely artificial fluorescent beta-barrel protein. As described by one of the main authors, Anastassia, in this forum post:
The paper presents many “firsts” in computational protein design. It is the first de novo design of the beta-barrel fold (one of the most described folds in the past 35 years, yet mysterious until now). It is also the first de novo design of a protein tailored to bind a small-molecule, which requires very high accuracy in the placement of side chains on protein backbones assembled from scratch. Additionally, we could show that these new proteins could fold and function as expected in vivo! We hope that the advances described in the paper will further enable the de novo design of many biosensors and catalysts tailored for specific applications.
Thanks to all the Rosetta@home volunteers who contributed to the validation of our designed proteins and binding sites.
Here is the link to the IPD webpage that contains a copy of the paper. The work was also featured in the news articles below (the news in Science contains a video of one of our proteins glowing in living cells).
https://www.bakerlab.org/index.php/2018/09/12/de-novo-fluorescent-proteins
https://cen.acs.org/physical-chemistry/periodic-table/Designer-protein-tackles-binding/96/i37
http://www.sciencemag.org/news/2018/09/watch-these-new-designer-proteins-light-when-they-hit-their-target
17 Sep 2018, 23:14:21 UTC
· Discuss
Congrats to the collaborative WeFold group for their recent paper published in Nature Scientific Reports. Thank you to all R@h volunteers who contributed to this work.
Congrats to the WeFold group for their recent publication, An analysis and evaluation of the WeFold collaborative for protein structure prediction and its pipelines in CASP11 and CASP12, in Nature Scientific Reports. As CASP13 is currently in full swing, this article describes the results and analysis of the CASP11 and CASP12 WeFold coopetition (cooperation and competition) . Most of the models used by WeFold for CASP12 (and currently for CASP13) were generated by Rosetta@home volunteers. Congrats and Thank you!
3 Jul 2018, 0:13:42 UTC
· Discuss
Rosetta Android version 4.10 released
This version uses a relatively recent version of the Rosetta source. It includes updates to the cyclic peptide folding protocol among other code updates and additions since the previous build. Please post comments and issues in this thread.
27 Apr 2018, 23:29:47 UTC
· Discuss
Science opinion article about protein engineering and David Baker
Checkout a recent Bloomberg science opinion article about some of the science behind this project, protein engineering, and David Baker, titled Protein Engineering May Be the Future of Science.
28 Mar 2018, 17:51:52 UTC
· Discuss
Rosetta 4.07 released
This version contains a bug fix for the cyclic peptide folding protocol. Please post any issues/bugs regarding this application in this thread.
27 Feb 2018, 18:33:12 UTC
· Discuss
Charity Event 2018
The Charity Team has chosen Rosetta@home for their 10th annual event to receive extended computation support for 2 weeks (January 14th 2018, 0.01h to January 27th 2018, 23.59h UTC). To participate, BOINC members are asked to leave their ‘Home Teams‘ and join the Charity Team to crunch Rosetta@home together without the normal ‘Race Conditions‘ during this time frame.
To join the Charity Team at Rosetta@home please click here:
https://boinc.bakerlab.org/rosetta/team_display.php?teamid=10787
You can find more information on the Charity Event forums:
http://forum.charity.boinc-af.org/index.php?board=12.0
and about the Charity Team: https://www.seti-germany.de/wiki/Charity_Team
Stats are also available at:
Team Stats (BoincStats): https://boincstats.com/en/stats/14/team/detail/10787/lastDays
User Stats (Sébastien): http://statseb.boinc-af.org/charity/
Comparison Stats (XSmeagolX): https://timo-schneider.de/sgstats/charity2018
Many thanks to the Charity Team for choosing Rosetta@home for their annual event!

4 Jan 2018, 0:30:08 UTC
· Discuss
The New York Times recently published an article about David Baker and Rosetta
Scientists Are Designing Artisanal Proteins for Your Body
The human body makes tens of thousands of cellular proteins, each for a particular task. Now researchers have learned to create custom versions not found in nature....
2 Jan 2018, 19:31:43 UTC
· Discuss
Recent Science and Nature publications. Congrats!
Two research publications were released last week in Science and Nature. The Science publication describes work which relied on computations from Rosetta@home most of which were from Android devices. The Nature publication did not directly use Rosetta@home due to the large size of the designs but used Rosetta. Congrats and thank you for your contributions!
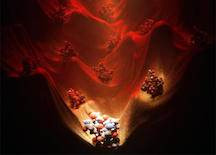
Comprehensive computational design of ordered peptide macrocycles. As described in the abstract, macrocyclic peptides composed of l- and d-amino acids were designed by near-exhaustive backbone sampling followed by sequence design and energy landscape calculations. More than 200 designs were predicted to fold into single stable structures, many times more than the number of currently available unbound peptide macrocycle structures. Nuclear magnetic resonance structures of 9 of 12 designed 7- to 10-residue macrocycles, and three 11- to 14-residue bicyclic designs, are close to the computational models. The results provide a nearly complete coverage of the rich space of structures possible for short peptide macrocycles and vastly increase the available starting scaffolds for both rational drug design and library selection methods.
Read more from UW Medicine News.
Evolution of a designed protein assembly encapsulating its own RNA genome. As described in the abstract, synthetic nucleocapsids composed of icosahedral protein assemblies with positively charged inner surfaces were computationally designed. The ability of these nucleocapsids to evolve virus-like properties by generating diversified populations and selecting for improved genome packaging and fitness against nuclease challenge was also explored. The results show that there are simple evolutionary paths through which protein assemblies can acquire virus-like genome packaging and protection. Considerable effort has been directed at ‘top-down’ modification of viruses to be safe and effective for drug delivery and vaccine applications; the ability to design synthetic nanomaterials computationally and to optimize them through evolution now enables a complementary ‘bottom-up’ approach with considerable advantages in programmability and control.
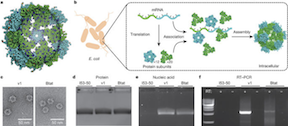
18 Dec 2017, 19:20:52 UTC
· Discuss
Research of the Year!
Congrats and thank you! Chemical and Engineering News has highlighted our work for research of the year. Your contributions provided much of the computing to make this possible. Thank you!
Read more.
14 Dec 2017, 19:24:28 UTC
· Discuss
Our rosetta and android apps have been updated to version 4.06
This app version was built using the latest Rosetta source. Please report issues in this thread.
21 Nov 2017, 20:14:17 UTC
· Discuss
Rosetta@home has contributed to a number of recent publications. Congratulations and thank you!
In Nature: building 20,000 new drug candidates. New de novo designed "mini-protein" binders were custom built to target either a deadly virus or a potent toxin and were shown to afford protection to mice. Read more.

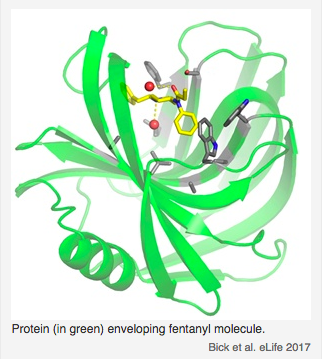
Sensors for the potent opioid fentanyl. Using a fully-automated Rosetta design pipeline, high-affinity fentanyl sensors capable of detecting environmental fentanyl were produced. Read more.
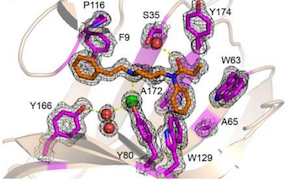
In Science: data-driven protein design. This work achieves the long-standing goal of a tight feedback cycle between computation and experiment and has the potential to transform computational protein design into a data-driven science. Rather than observing thousands of complex natural proteins to try to deduce their folding rules, over 15,000 new, simpler proteins were built – all designed using Rosetta. Through multiple design rounds, features that led to successful folding were learned and incorporated into the design pipeline. Read more.

5 Oct 2017, 18:42:22 UTC
· Discuss
The rosetta application has been updated to 4.04 for Windows platforms
The "rosetta" application has been updated to 4.04 for Windows platforms to address the Windows BOINC client 7.8.2 issues. This is a relatively recent version of the Rosetta software source and will eventually replace the "minirosetta" application. Please report issues/bugs in this thread.
5 Oct 2017, 17:43:12 UTC
· Discuss
The minirosetta application has been updated to 3.78. This version includes a patch to prevent fatal errors when using the Windows BOINC client version 7.8.2. There are known issues with this client version for Windows platforms and the BOINC developers are working on a fix. To report bugs, go to this thread.
3 Oct 2017, 18:19:20 UTC
· Discuss
Welcome to our updated website!
After many years of service, we are happy to announce that our old website and servers have been retired and we have finally released our new website powered by the latest BOINC software and new hardware. Everything is new and modernized.
If you have any comments or would like to report any issues regarding the new website please post your feedback in this thread.
23 Jun 2017, 4:17:54 UTC
· Discuss
Outage notice
I'm pretty sure this message never appeared at the datetime stated. First I knew was 36 hours later and I'm pretty sure I logged on 24 hours after this notice.
Still, can't be long now. I note the upload server isn't back quite yet.
20 Jun 2017, 0:00:00 UTC
· Discuss
Journal post from David Baker
Good morning everyone... 😆 It is with great pleasure that I have been helping this project, but lately I see that it is a little abandoned. Many people stopped participating, and the work has grown a lot. Please help, because from what I have read, our help has been very important for the discovery of new proteins that help a lot in the development of therapies to cure diseases. I appeal to users to try a little harder. Thank you all.
17 Jun 2016, 0:00:00 UTC
· Discuss
The minirosetta application has been updated to 3.65
Thanks you sharing
12 Oct 2015, 0:00:00 UTC
· Discuss
Rosetta Graphics released for Windows
cool,
27 Nov 2005, 0:00:00 UTC
· Discuss
Archived news.
Our Android application has been updated to version 3.83. This update includes bug fixes and new protocols (specifically for cyclic peptide folding and design), and should be significantly more stable for Android 4 to 7.+ platforms. Please report bugs in this thread.
3 Apr 2017, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
NOVA recently featured some of the work that all of you are contributing to:
http://www.pbs.org/wgbh/nova/physics/origami-revolution.html (the 8 minute segment on our work starts at 20:30)
The Economist also had an article on the work you are contributing to:
http://www.economist.com/news/science-and-technology/21716603-only-quarter-known-protein-structures-are-human-how-determine-proteins
Your contributions are highlighted particularly clearly in the Geekwire article:
http://www.geekwire.com/2017/big-data-rosetta-protein-puzzles
which is titled "Big data (and volunteers) help scientists solve hundreds of protein puzzles"
27 Feb 2017, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
Hi Everybody!
this has been a good week with papers in this and last weeks Science magazine. your contributions were essential for both breakthroughs! here is some of the press you might find interesting:
http://www.geekwire.com/2017/big-data-rosetta-protein-puzzles
https://www.theatlantic.com/science/archive/2017/01/unravelling-lifes-origami/513638
http://www.geekwire.com/2017/uw-protein-pockets
thank you again for your invaluable contributions to this research!!
David
20 Jan 2017, 0:00:00 UTC
· Discuss
Archived news.
More good news!
Our paper titled "Protein structure determination using metagenome sequence data" was released today in the journal Science. We would like to thank all Rosetta@Home participants who provided the computing required for this work. In the paper, we describe using predicted co-evolving contacts from metagenomics sequence data and Rosetta to accurately predict the structures for 622 protein families that are not represented in the PDB. Among these structures, over 100 were new folds. Since experimental protein structure determination is costly and often difficult, this study highlights the ability to use computational methods with metagenomics data for reliably structure determination. With the rapidly growing size of genomics data, the future in mapping the structure space of protein families looks bright! Thank you Rosetta@Home participants!
Here is an interesting perspective written by Johannes Söding about the paper and it's significance, "Big-data approaches to protein structure prediction".
and related news articles:
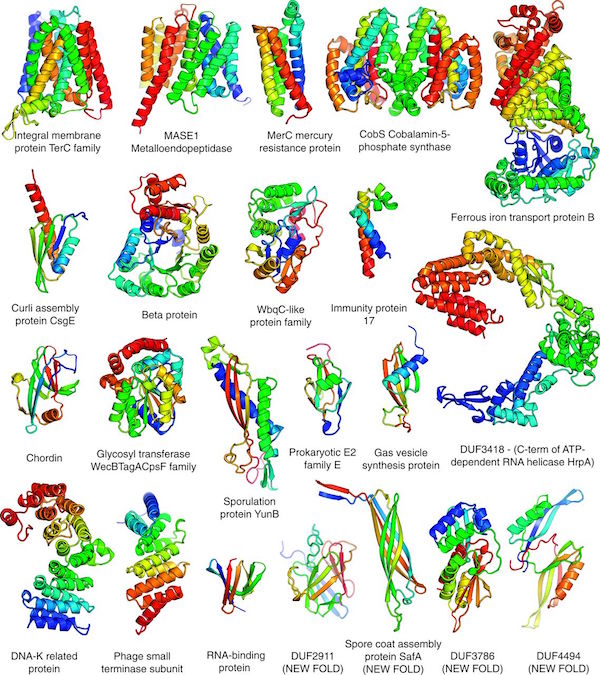
19 Jan 2017, 0:00:00 UTC
· Discuss
Archived news.
Happy New Year!
As many of you are likely aware of, we had an outage yesterday that we are just recently recovering from. The University of Washington campus and surrounding Seattle area had a power outage that lasted for a couple hours. For more info about the outage click here. It may take a day or so to get back to normal operation. Sorry for any inconvenience.
9 Jan 2017, 0:00:00 UTC
· Discuss
Archived news.
Some good news!
We recently published an article in Nature titled "Accurate de novo design of hyperstable constrained peptides". We would like to thank all Rosetta@Home participants for their help with this work. In the paper, we present computational methods for designing small stapled peptides with exceptional stabilities. These methods and designed peptides provide a platform for rational design of new peptide-based therapeutics. Constrained (stapled) peptides combine the stability of conventional small-molecule drugs with the selectivity and potency of antibody therapeutics. The ability to precisely design these peptides in custom shapes and sizes opens up possibilities for "on-demand" design of peptide-based therapeutics.
Other developments described in the paper:
We are now working to use these computational methods for designing peptides that target therapeutically relevant targets, such as, enzymes that impart antibiotic resistance in pathogenic bacteria.
Structure prediction runs on Rosetta@Home for these designed peptide models played a key role in selection of good designs that were experimentally synthesized and characterized. Thank you all for your help in making this work possible! -- Gaurav B.
23 Sep 2016, 0:00:00 UTC
· Discuss
Archived news.
In the last few weeks our project has experienced significant issues resulting in slower than usual work unit distribution, result processing, and credit granting. The cause of this was due to the increasingly large number of new hosts causing our database server to become very sluggish and eventually run out of disk space. Our short term solution was to reconfigure and optimize the project configuration and existing database server, purge old data quicker than usual, and temporarily stop resource intense database queries. This recovery mode will continue until the project stabilizes which may take a few days to a week.
An interim solution will be to temporarily upgrade our database server and thanks to our sys admins, we already have a machine ready to go that has plenty of disk space and double the memory. The upgrade will require a day of downtime which is planned to happen early this week.
The long term solution will be a complete system hardware upgrade to all our servers. The BOINC server software will also be upgraded. We are in the process of ordering these machines and hope to have them running within the next few months.
The project is somewhat stable now and clients should be getting work as usual. However, result processing and credit granting may still be slow and our status information/page may not be up to date as we are in recovery mode and our servers continue to catch up on things. Work history will also be shortened to temporarily save space. We expect things to be back to normal in a few days to a week.
Sorry for any inconvenience and thank you for your continued contributions!
12 Sep 2016, 0:00:00 UTC
· Discuss
Archived news.
We've had an unplanned project crash. The project is back online but you might experience interruptions while the system catches up.... -KEL [Sun Sep 4 07:53:59 PDT 2016]
4 Sep 2016, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
We all collectively made the cover of the July 22nd Science-check it out! This same issue also has a news feature on our work, and an article on designed icosohedral cages made from two different designed protein building blocks. For more information and links to the papers, see Ratika's post in her thread.
1 Aug 2016, 0:00:00 UTC
· Discuss
Archived news.
Another paper from the Baker lab in Science! Designed Protein Containers Push Bioengineering Boundaries.
"In this paper, former Baker lab graduate student Jacob Bale, Ph.D. and collaborators describe the computational design and experimental characterization of ten two-component protein complexes that self-assemble into nanocages with atomic-level accuracy. These nanocages are the largest designed proteins to date with molecular weights of 1.8-2.8 megadaltons and diameters comparable to small viral capsids. The structures have been confirmed by X-ray crystallography. The advantage of a multi-component protein complex is the ability to control assembly by mixing individually prepared subunits. The authors show that in vitro mixing of the designed subunits occurs rapidly and enables controlled packaging of negatively charged GFP by introducing positive charges on the interior surfaces of the two copmonents.
The ability to design, with atomic-level precision, these large protein nanostructures that can encapsulate biologically relevant cargo and that can be genetically modified with various functionalities opens up exciting new opportunities for targeted drug delivery and vaccine design." - from the IPD website.
More information can be found at this link.
Thank you all for your contributions!
29 Jul 2016, 0:00:00 UTC
· Discuss
Archived news.
A brand new paper from the Baker lab in Nature! Thank you yet again for your contributions, Rosetta@home volunteers. Although R@h was not directly used for these designs due to size and memory limitations, your contributions are instrumental in Rosetta development enabling such science. Thank you!
"Design of a hyperstable 60-subunit protein icosahedron" describes the design of a 20-sided nano-sized particle that could be used to deliver drugs or to develop powerful new vaccines.
Author and Baker lab graduate student Yang Hsia spoke on a Nature podcast on the work. Have a listen to hear about this awesome work from the protein design experts themselves! Links below.
Paper
Podcast Under 16 June 2016 "Protein football"

17 Jun 2016, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
I recommend listening to graduate student Yang Hsia's podcast on designing protein footballs which you can find on the Nature web site this week to go along with his paper. (see Ratika's post for the link if you can't find it). Thank you all again for all of your contributions to our research-we couldn't do it without you!
17 Jun 2016, 0:00:00 UTC
· Discuss
Archived news.
We've come out with a breakthrough paper in Science titled 'De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity'.
This is an exciting and significant breakthrough for de novo protein design. A particular challenge for current protein design methods has been the accurate design of polar binding sites or polar binding interfaces, both of which require hydrogen bonding interactions. Hydrogen bond networks are governed by complex physics and energetic coupling, that until now, could not be computed within the scope of design. The computational method described in this paper, HBNet, now provides a general method to accurately design in hydrogen bond networks. This new capacity should be useful in the design of new enzymes, proteins that bind small molecules, and polar protein interfaces. Thanks Rosetta@home community for your participation and help!
An article on this work was also published in Geekwire http://www.geekwire.com/2016/uw-researchers-add-new-twists-protein-designs.
10 May 2016, 0:00:00 UTC
· Discuss
Archived news.
We'll be down for an hour or so for some maintenance. Sorry for any inconvenience.
28 Apr 2016, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to 3.73. This version includes new protocols and a fix for an improved score function. To report bugs, go to this thread.
31 Mar 2016, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
The results on the flu neutralizing protein you helped us design have now been published. You can get the paper, like all of our papers, from our lab web site, and read what journalists are saying at
http://cen.acs.org/articles/94/i6/Designer-Protein-Promising-Antiflu-Agent.html
12 Feb 2016, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to 3.71. This version includes an improved score function, new protocols, and slightly updated graphics. To report bugs, go to this thread.
20 Jan 2015, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
We just published a paper in Nature that depended critically on all of your contributions! You can get the pdf for this paper on our website; you can also get pdfs of the many other papers we've published in the last couple of years that relied heavily on rosetta@home.
18 Jan 2016, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to 3.65. To report bugs, go to this thread.
12 Oct 2015, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to 3.62. To report bugs, go to this thread.
3 Sep 2015, 0:00:00 UTC
· Discuss
Archived news.
We are experiencing technical difficulties with our servers and are currently troubleshooting. We hope to have things back to normal soon. Sorry for any inconvenience.
31 Aug 2015, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to 3.59. To report bugs, go to this thread.
15 Jun 2015, 0:00:00 UTC
· Discuss
Archived news.
Here is a video of David Baker describing some of the research going on in the lab from his recent talk at the American Association for the Advancement of Science (AAAS) meeting. Note: you may have to scroll down a little to view the video.
24 Mar 2015, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to 3.54. To report bugs, go to this thread.
9 Mar 2015, 0:00:00 UTC
· Discuss
Archived news.
Happy New Year to all! We just posted a brief synopsis of our CASP11 results in this thread post, and would like to say thank you for all your contributions! Thank you all!
6 Jan 2015, 0:00:00 UTC
· Discuss
Archived news.
Here's an update from David Baker on the Institute for Protein Design's progress: Letter from the Director – IPD Update
15 Aug 2014, 0:00:00 UTC
· Discuss
Archived news.
Slowdown Update: As it turns out, the slow-down experienced during the last week of July was the result of a very large surge in users joining the project through a new campaign by http://charityengine.com. As the servers were behaving properly - just overwhelmed - and DK was on a well-deserved holiday, it was difficult for the rest of us to pin-point the cause. You can see the recent surge here.
We are looking at changing/expanding our webserver frontend to be more resilient to surges like this in the future. Yet again, we apologize for the frustration caused. -KEL
5 Aug 2014, 0:00:00 UTC
· Discuss
Archived news.
We are aware of significant network slow-down between the subnet upon which our servers sit and the Internet beyond the UW campus. We are working with the UW's Network Operations team to pinpoint the cause. We apologize for the frustration caused. -KEL
30 Jul 2014, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
We have recently made great progress in designing self assembling materials from proteins and in designing proteins which selectively kill tumor cells. These efforts have benefited tremendously from Rosetta@Home! News releases on these advances are at
http://hsnewsbeat.uw.edu/story/self-assembling-nanomachines-start-click
http://hsnewsbeat.uw.edu/story/computer-designed-protein-causes-cancer-cells'-death
20 Jun 2014, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to 3.52. This version includes a couple bug fixes for symmetric modeling. To report bugs, go to this thread.
28 May 2014, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to 3.50. For details and to report bugs, go to this thread.
29 Apr 2014, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
There has been very exciting recent progress in designing vaccines and small molecule binding proteins using Rosetta that is described in two recent papers in Nature. These and other recent advances are described in the new Rosetta@Home Research Updates thread. It was suggested there that we send out a monthly email newsletter describing recent progress--we haven't done this before to avoid clogging everybody's inboxes but we certainly could if there is interest.
2 Apr 2014, 0:00:00 UTC
· Discuss
Archived news.
UPDATE - We successfully moved all the hardware that supports Rosetta@Home to new a home, some 25 m from it's old location. We did this to help balance the heat load in the datacenter as well as gain more physical space and electrical power for future R@H expansion (if we can afford it). -KEL and DOVA
5 Feb 2014, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
An article mentioning Rosetta@Home recently appeared in the Globe and Mail:
http://www.theglobeandmail.com/news/national/meet-two-pioneers-of-immune-research/article15101089/
10 Nov 2013, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to 3.48. Please report bugs in this thread
10 Oct 2013, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
Our new Institute for Protein Design is making considerable progress as you can see at http://ipd.uw.edu. we are now using Rosetta@home to rigorously evaluate all computer-based designs before we create synthetic genes to make the proteins in the laboratory. this is dramatically increasing our success rate at designing proteins with new functions. your contributions continue to be absolutely invaluable!
25 Sep 2013, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
We have discovered how to make several new classes of protein structures! Rosetta@home has been absolutely critical in this work: when we design a sequence to fold into a new structure, the last thing we do before ordering a synthetic gene so we can make the protein in the laboratory is to send it out to you to predict the structure-if it folds to the structure we designed, we go ahead with it, but if you find that the lowest energy state is a different structure we go back to the drawing board. Our success rate in making brand new structures is far higher than I or anybody else ever expected, and the reason the success rate is so high is that your calculations provide a very stringent test of whether the designed sequence will actually fold the way it is supposed to. In the next few weeks I and other scientists here will describe to you the new classes of proteins we are making, and the many applications they will be useful for. Thank you for your absolutely essential contributions to this newly emerging scientific field!
20 May 2013, 0:00:00 UTC
· Discuss
Archived news.
We again experienced networking problems with the backend of the R@H system. We're up and running again, but the system will be sluggish as the backlogged tasks are worked through. -DOVA and KEL [Fri May 17 13:55:23 PDT 2013]
17 May 2013, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application is updated to 3.46. For details and to report bugs, go to this thread
26 Apr 2013, 0:00:00 UTC
· Discuss
Archived news.
The backend network to R@H became 'fussy' over the past week resulting in an overall slowdown in performance (related to the power outage?). The network was 'rebooted' and things are back to normal. -KEL and DOVA
24 Apr 2013, 0:00:00 UTC
· Discuss
Archived news.
The University of Washington and the surrounding neighborhoods expereince a ~2 hour power failure late last night. While the R@H gear is located the UW's datacetner on campus, the equipment is not protected by generator backup. Everything needed to be rebooted and reset up this AM. -KEL and DOVA
12 Apr 2013, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
We have learned to design a new class of proteins which could be useful both as drugs and in sensors. As I've explained before, most drugs are small molecules with fewer than 50 atoms. There are many drugs, such as blood thinning agents, which are very dangerous if given in too large doses. We have succeeded in designing proteins which bind to specific small molecules very tightly. These proteins could be used as antidotes in case of overdose with the target small molecule-for example we've designed a protein which could be useful to treat toxicity due to overdose of the drug digoxin used to treat heart disease. With collaborators, we are working to use these designed proteins to detect levels of the target small molecules in the blood or in the environment.
11 Mar 2013, 0:00:00 UTC
· Discuss
Archived news.
R@H will be down for a few hours starting at noon today in order to diagnose and fix an issue with our filesystem. Sorry for any inconvenience and thanks for your support!
21 Feb 2013, 0:00:00 UTC
· Discuss
Archived news.
One of the R@H servers stopped for a while and this caused quite a traffic jam for all the results coming back from all of you. We've brought things back online and the queue is slowly clearing. Sorry for the bother. R@H-IT
18 Feb 2013, 0:00:00 UTC
· Discuss
Archived news.
Our servers were down for about an hour today so that we could upgrade the fileserver for both Rosetta@HOME and our other project Robetta. Sorry for the interruption and thanks for your patience and your continued contribution to our research efforts! -KEL and DOVA
20 Jan 2013, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
The CASP10 meeting just finished and all the results are on line so you can see what all your contributions made possible! Ray has posted a great summary of the results in the CASP10 thread on the science message boards. Overall, Rosetta@home was top or near top in most categories. I hate to embarrass David Kim who created RosettaHome and has kept it going, but his contact guided predictions were the highlight of CASP10 as they were vastly better than those of any other group. You can see this at
http://predictioncenter.org/casp10/results.cgi?view=targets&model=all&tr_type=others&groups_id=4
David's predictions are the black lines, those of other groups are in orange, better models stay below the rest of the pack.
Thanks to all of you who contributed to CASP10!!
15 Dec 2012, 0:00:00 UTC
· Discuss
Archived news.
An application update to version 3.45 has been made to address a serious bug occurring on Windows 64 bit platforms. If you are running Windows 64 bit clients, please cancel existing Rosetta@home jobs that are either cached or running the older 3.43 application version. You may see a blank Rosetta@home screensaver window which you should manually exit/close. We are sorry for any inconvenience and we are updating our scripts and process to prevent this from happening again in the future. For more information about this issue you can go to this message board thread. Thank you to those that have posted on this thread and have helped to identify the root cause of this issue.
14 Nov 2012, 0:00:00 UTC
· Discuss
Archived news.
A number of users are having issues with our screensaver application automatically running in a pop-up window. We currently are not able to reproduce this on our test machines so we are asking users to please post any information that may help us track down this bug. Please post on this message board thread specific information about the boinc client version and platform version. We are also investigating another issue that may be related to an increase in our application database size.
14 Nov 2012, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@Home software updated to version 3.43
13 Nov 2012, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
We would like to acknowledge the Rosetta@Home participants who found the lowest energy structures for Nobu and Rie's designed proteins:
Fold-I : Aalelan (United States)
Fold-II : Jef (United States)
Fold-III : georgebg (Bulgaria)
Fold-IV: medvjet009(Czech Republic)
Fold-V : _2e_ Russia.
See Nobu's message board thread on "Principles for designing ideal protein structures" (https://boinc.bakerlab.org/rosetta/forum_thread.php?id=6113) for more information including a figure illustrating the critical role played by Rosetta@home in this work. And check out the latest on slashdot:
http://science.slashdot.org/story/12/11/09/020241/proteins-made-to-order
This and other publications from the lab are available at depts.washington.edu/bakerpg
Thanks again to everybody!!
10 Nov 2012, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
With your help, we have made an exciting breakthrough in protein design that is reported in a research article titled "Principles for designing ideal protein structures" in the journal Nature today. You can read about it at
http://www.nature.com/news/proteins-made-to-order-1.11767
In this paper, we describe general principles for creating new proteins from scratch. The new Institute for Protein Design is using these principles to design new proteins to treat disease.
Rosetta@home was absolutely critical to this work as described in the news article; Figure 3 in the paper shows how all of your contributions were used to test designed sequences to see if they folded up to the right structure. Most of the work units we are sending out on Rosetta@home these days are for exactly these kind of tests on the new proteins we are designing--this is absolutely critical to the research and to the development of new therapeutic and other functions. Thank you again for all of your contributions!
8 Nov 2012, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
I have exciting news. We and the University of Washington are starting up a new Institute for Protein Design to design new proteins to address current challenges in medicine, energy, and other areas. You can learn more about the institute at http://depts.washington.edu/ipd/. Rosetta@home is and will continue to be a critical part of our efforts. For every new potential protein therapeutic we design, we use Rosetta@home to test whether it will actually fold into the desired structure. And we need help! We have quite a backlog of exciting new designed proteins to test on Rosetta@home because we are designing proteins for quite a range of problems-new anti flu proteins, anti-cancer proteins, and new materials--and it takes 3000-5000 work units to test each one. This Rosetta@home testing is becoming the slow step in the whole design process, often taking over 10 days to complete. So please tell your friends and relations to join us!
A generous donor has provide funds which we want to use to invite 5-10 Rosetta@home participants to visit the Institute and see what we are trying to accomplish first hand. More on this in my next post.
21 Oct 2012, 0:00:00 UTC
· Discuss
Archived news.
Network Outages: We've just experienced a DNS outage due to a change-over in our Domain Registrar. Things should be back up and running now. We appreciate your patience. -KEL
4 Oct 2012, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
The native structures are slowly being released for CASP10 targets; all of them will be available by the end of November. In the meantime, you can look at the results of a much larger scale test of prediction methods called CAMEO. CAMEO takes newly solved protein structure before they are published, and sends the amino acid sequences out to structure prediction servers. This happens every week, so it is great to assess new methods as they are being developed. You can look at the results, as well as get more information about CAMEO, at
http://beta.cameo3d.org/modeling/weekly_summary.html
The best number to compare is the "Average accuracy (all targets)" as some servers only model the easy ones. The good thing about CAMEO is that there are many more test cases than CASP, and also that results are released each week so we can see what is working well and what needs to be improved. You will see that ROBETTA, which is now using some of your computing resources, is doing pretty well recently; before this we had problems with some targets not getting enough work units before the server deadline.
28 Sep 2012, 0:00:00 UTC
· Discuss
Archived news.
Network Outages: As part of the UW's continuing datacenter consolidation, the network topology upon which Rosetta@home is run was changed yesterday. Since that time we've been shaking out the various hiccups that result from changing things in such a busy system. We, the IT crew, apologize for the troubles and will try to get them ironed out as soon as we can. We appreciate your patience and your continued contributions to our research efforts. -KEL
28 Sep 2012, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
We are now testing the latest batch of novel designed proteins that you have helped us create in our brand new Molecular Engineering laboratory at the UW. You can see pictures of the space where the rosetta@home computed designs are being experimentally tested in a recent newspaper article:
http://seattletimes.com/html/localnews/2019068219_molecularlab05m.html
10 Sep 2012, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
A recent paper in Nature Biotechnology describes how we have combined computational protein design with the high throughput DNA sequencing methods developed for sequencing the human genome to generate potent influenza virus inhibitors. These designed proteins block infection by the flu virus in cell culture experiments, and they are now going through the (quite lengthy) process of being developed as possible anti-flu drugs.
4 Sep 2012, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@Home software updated to version 3.41.
24 Aug 2012, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
Many common materials such as silk and wool are made out of regular repeating arrays of proteins, and symmetric protein arrays make up the coats of viruses and many other assemblies inside cells. The ability to robustly design self assembling materials made out of proteins would have huge numbers of applications-the naturally occurring assemblies are useful, but imagine if we could make custom materials for 21st century problems. In this weeks Science magazine, we describe the use of Rosetta to design self assembling protein nano structures with very high accuracy. We are now attempting to create many different types of symmetric materials, and you will be seeing more symmetric calculations on your rosetta@home screen server. Thank you for making possible this completely new approach to nanotechnology!
10 Jun 2012, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@Home software updated to version 3.31. Addresses issues with improper initialization of inputs in hybrid protocol for comparative modeling.
16 May 2012, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
A big THANK YOU to all of you who have scaled up your contributions to Rosetta@Home-this is a record level of computing power for us and is super well timed. THANKS!!!
8 May 2012, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
I have just been told the very good news that Rosetta@home will be the first project of the BOINC pentathlon, and would like to thank all of the participating teams. I also just learned from the discussion thread that Rosetta@home will be the project of the month for BOINC synergy-this is more excellent news!!
Your increased contributions to rosetta@home could not come at a better time! We've been testing our improved structure prediction methodology in a recently started challenge called CAMEO. For most of the targets, the Rosetta@home models are extremely good, but for a minority of targets the predictions are not good at all. We've now tracked down the source of these failures and it is what we are calling "workunit starvation"; in the limited amount of time the Rosetta server has to produce models (2-3 days) in these cases very few models were made-this happens because many targets are being run on the server so that only a fraction of your cpu power is focused on any one target. while we are working to fix this internally, by far the best solution is to have more total CPU throughput so each target gets more models.
You can follow how we are doing at http://www.cameo3d.org/. You will see that Robetta is one of the few servers whose name is not kept secret-this is because Rosetta is a public project. Our server receives targets from CAMEO and soon CASP, sends the required calculations out to your computers through Rosetta@home, and then processes the returned results and submits the lowest energy models.
We are excited that the workunit starvation problem may go away through your increased efforts for Rosetta@home. Thanks!!!
29 Apr 2012, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@Home software updated to version 3.30. Addresses issues with Cartesian Relax and the modeling of disulfides in the hybrid protocol for comparative modeling.
27 Apr 2012, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
I've described in the past our work using Rosetta and Rosetta@Home to create new enzyme catalysts. In Nature Chemical Biology last month we describe the design of an enzyme which destroys organophosphate
nerve agents and pesticides. These compounds kill by blocking key enzymes, and our designed enzyme eliminates this toxicity. This illustrates how Rosetta@Home enzyme design work can help to solve current problems, including man-made problems.
8 Apr 2012, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@Home software updated to version 3.26. This update includes several enhancements for symmetry in the hybrid protocol for comparative modeling. If you encounter any issues, please let us know here
5 Apr 2012, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
In the last two months we believe we have made quite a breakthrough in structure prediction, and are excited to test the new method in CASP10. We need your help though--we are now testing many aspects of the new approach and are seriously limited by available CPU cycles. There are now so many flu inhibitor design and structure prediction jobs queued up on Rosetta@Home that there is an eight day wait before they are getting sent out to you. This would be a great time to temporarily increase Rosetta@Home's share on your computers and/or recruit new users--we need all the help we can get! thanks! David
30 Mar 2012, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@Home software updated to version 3.24. This update includes support for symmetry in the hybrid protocol for comparative modeling. If you encounter any issues, please let us know here
14 Mar 2012, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@Home software updated to version 3.22.
13 Feb 2012, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
Last year we described in Science magazine the design of a new enzyme which catalyzes a chemical reaction called the Diels Alder reaction involving the formation of two carbon-carbon bonds. This reaction is interesting because no natural enzymes are known to catalyze the reaction. However, it wasn't a very good enzyme, and we asked FoldIt players to try to improve it. As described in Nature Biotechnology this month, remarkably FoldIt players were able to make the designed enzyme 20 times faster by inserting a completely new loop which helps the enzyme bind the chemicals it links together. The combination of Rosetta@Home and FoldIt is turning out to be powerful indeed for solving challenging problems in biomedicine!
29 Jan 2012, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
In response to requests from many of you, we will be posting descriptions of the many scientific problems currently being tackled with Rosetta@Home on the Science message boards in the next couple of weeks--stay tuned! I also want to describe a new research direction we are now embarking on aimed at future cancer therapies. There are a small set of proteins which are frequently found at much higher levels than normal on the surface of cancer cells. We are starting to design small proteins which bind tightly to these tumor cell markers. If we are successful, we have collaborators who will be testing these proteins for their ability to target cancer cell killing agents to the tumors.
16 Jan 2012, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@Home software updated to version 3.20. This should fix the graphical issues some users were seeing with version 3.19.
13 Jan 2012, 0:00:00 UTC
· Discuss
Archived news.
The hardware hosting the Rosetta@Home project is being moved from one datacenter to another on the UW campus. We are using this disruption to update the sagging, aging gear that runs the project. All of this will result in a few days of down time. We will work to keep the outage to a minimum, but you migh want to grab enough work for a 4-5 day period. We appreciate your patience, interest and continued contributions to our research. -KEL
2 Jan 2012, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@Home software updated to version 3.19
19 Dec 2011, 0:00:00 UTC
· Discuss
Archived news.
David Baker receives UW Medicine's Inventor of the Year award.
From the award announcement " the Inventor of the Year award is given to individuals who have translated research from the bench, through partnerships with the biomedical industry, to a product or process that has had a major impact on healthcare and the local economy" . Read more...
26 Oct 2011, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@Home software updated to version 3.17
26 Oct 2011, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
Today's issue of Science magazine describes an exciting new approach to HIV vaccine design using Rosetta. In contrast with other viruses such as polio and influenza, inactivated HIV or HIV proteins have not worked as vaccines, and hence as you know there is currently no effective HIV vaccine. Our approach to vaccine design is to take the bits of the HIV surface protein that people make antibodies to, and using Rosetta graft them onto small stable scaffolds that can be made in large quantities and potentially could be useful as vaccines. We've shown earlier that this can be done straightforwardly with Rosetta if the bits of the HIV protein are contiguous along the sequence, but it is much harder if the antibody recognizes multiple bits close in three dimensions but far in sequence. In this paper we show how such "discontinuous" epitipes can be transferred from HIV gp120 to a simple scaffold protein. More work will be required to determine whether this or other vaccine candidates designed using this approach will be effective as HIV vaccines-let us all hope so!!
22 Oct 2011, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
A recent issue of Nature describes an exciting result from Rosetta@home in collaboration with the NMR spectroscopy laboratory of Lewis Kay in Toronto. Like almost all machines, proteins in order to carry out their functions have to move (change their conformation somewhat) but it has been extremely difficult to determine what these conformational changes are. Lewis Kay's group has developed new methods for getting experimental information on the higher energy very shortlived conformations proteins visit while carrying out their functions. This data is not sufficient to determine the structure of these "excited state" conformations using conventional methods. However, as the paper shows, we can use these experimental data to guide Rosetta and Rosetta@home structure calculations, and produce models of these states. We went one step further than this in the paper by using Rosetta design calculations to stabilize the excited state, and subsequent experiments confirmed the validity of the model. This combination of experimental NMR data, Rosetta structure calculations, and Rosetta design should be very powerful in understanding how proteins carry out their functions.
6 Oct 2011, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
Today's issue of Nature Structural Biology reports the determination of the structure of a protein by FoldIt players. This is exciting because it is perhaps the first example of a long standing scientific problem solved by non-scientists. You might read about this in your newspaper; here is a report that does a good job in explaining how FoldIt came out of Rosetta@home:
http://the-scientist.com/2011/09/18/public-solves-protein-structure/
19 Sep 2011, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
This week's issue of Nature magazine has an exciting article (http://www.nature.com/nature/journal/vaop/ncurrent/full/nature10154.html) describing work we are doing with collaborators using Rosetta to design a new class of inhibitors of amyloid fibril formation. Amyloid fibrils have been implicated in Alzheimer's and many other diseases. The designed peptides are not suitable for use as actual therapeutics in their present form, but hopefully will help lead the way to effective drugs.
18 Jun 2011, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 3.14. For details and to report bugs, go to this thread.
15 Jun 2011, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
A recent issue of Nature describes an exciting approach we are taking with collaborators to fight Malaria. The title of the paper is "A synthetic homing endonuclease-based gene drive system in the human malaria mosquito" and the PDF is available at my lab web site. The idea is to use enzymes which cut within critical genes in mosquitos to greatly reduce the number of malaria parasite infected mosquitos. There are still many issues that must be overcome for this strategy to be used against malaria in the real world, but this paper is an important first proof of concept of the strategy.
18 May 2011, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
This week's issue of Science magazine features an article on the use of Rosetta@Home to design novel proteins which bind tightly to the Spanish Flu (H1N1) Influenza Virus. The paper shows that the experimentally determined atomic structure of the complex between one of the designed proteins and the virus is precisely as in the computer model. The designed proteins block the function of the flu surface protein in biochemical tests, and we are guardedly optimistic that the designs will block flu infection. This is an important milestone for computational protein design (and for distributed computing)--the first atomic level accuracy design of a high affinity protein-protein interface, and the designed proteins are exciting leads for new flu therapeutics. In the next few months, we will be using Rosetta@Home to design proteins that bind tightly and hopefully block other pathogens which cause disease. Thanks to all Rosetta@home users for their invaluable contributions to this research!!
14 May 2011, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
Graduate student Shawn Yu is now posting on current Rosetta@home efforts to design inhibitors for viruses that cause disease in the "Design of Protein Interactions" thread on the Science message boards. Take a look if you are interested; he is happy to answer questions in the thread as well.
7 May 2011, 0:00:00 UTC
· Discuss
Archived news.
Journal post from David Baker
The paper on the Rosetta method which allows determination of the structures of a large class of proteins using limited crystallography data has now been published in Nature magazine. Thanks to all of you for making this work possible!
4 May 2011, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: We are going to update our scheduler tomorrow, Thursday the 24th. The project will be offline intermittantly throughout the day.
23 Feb 2011, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: We will be offline for a brief 1-2 hour period tomorrow, Thursday the 27th, starting at around 10am PST for maintenance.
26 Jan 2011, 0:00:00 UTC
· Discuss
Archived news.
As many of you are probably aware of already, we've been working hard to try to resurrect the project with the resources we have at hand. Today we replaced a very small temporary disk with a larger 3TB one that we received from the vendor a few days ago. With this disk in place we should soon be back up and running. We have yet to restore data from jobs before the disk failure so these older jobs will still be pending. However, all pending jobs will eventually be granted credit. The good news is that the data from the failed disk has been recovered and we are currently copying it to the new disk. Thank you all for your patience.
13 Jan 2011, 0:00:00 UTC
· Discuss
Archived news.
Well, our luck ran out. The SAN controller that has been causing so much trouble in the last few months finally tipped over in a rather distructive fashion, corrupting the binary tree on which the filesystem is based. We're trying to rebuild the thing but the sheer number of files in the filesystem (> 10M files) makes this process very, very slow. We're bringing the project up from a recent backup (12/09/10) but the backup wasn't a perfect replica of the environment, so we're having to scramble to get all the parts working together again. We only need a few more weeks and then our new, next generation SAN will be ready to be put into place... I just thought the old one would last a few more week. I apologize for the hassle and appreciate your patience as we get things online again... KEL 01/07/11
7 Jan 2011, 0:00:00 UTC
· Discuss
Archived news.
Today's heros are Keith and Darwin, our systems administrators and hardware architects. Yesterday, our main filesystem crashed hard. There were warning lights flashing behind every disk on the SAN and it looked pretty grim. Thankfully, Keith and Darwin were able to pinpoint the problem to two redundant laser modules for the fiber optic loops (it was amazing and unlucky that both failed). The laser modules have since been replaced and the filesystem is back up. We'll be starting up the project again shortly. Thanks for your patience.
9 Dec 2010, 0:00:00 UTC
· Discuss
Archived news.
Read about recent work using Rosetta to design flu virus inhibitors, HIV vaccines, and HIV destroying nucleases in David Baker's journal and the Design of protein-protein interfaces thread.
23 Nov 2010, 0:00:00 UTC
· Discuss
Archived news.
Follow Rosetta@HOME on Twitter! @rosettaathome We will be posting R@H specific scientific updates and interesting research news there!
9 Nov 2010, 0:00:00 UTC
· Discuss
Archived news.
We're back online, thanks!
5 Nov 2010, 0:00:00 UTC
· Discuss
Archived news.
The project is temporarily down for maintenance. It should be back up in an hour or so. Sorry for any inconvenience.
5 Nov 2010, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 2.17. For details and to report bugs, go to this thread.
31 Oct 2010, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 2.16. For details and to report bugs, go to this thread.
9 Oct 2010, 0:00:00 UTC
· Discuss
Archived news.
A few days ago our main filesystem went down but we are finally back to normal operation. Sorry for any inconvenience.
1 Oct 2010, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 2.15. For details and to report bugs, go to this thread.
21 Sep 2010, 0:00:00 UTC
· Discuss
Archived news.
As many of you have already noticed, we are experiencing some issues with our work unit generators. We are working on a fix right now and will hopefully be back up to speed soon. Thanks for your patience.
1 Sep 2010, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home related advances have been featured in the national media twice in the last month. The first advance was the design of a protein catalyst that joins two molecules in a way not found in Nature; this was reported in Science magazine in July. The second advance is the finding that FoldIt players can solve hard structure prediction problems; this was reported in Nature magazine this week. These advances were covered in the New York Times, the LA times, and likely in your local newspaper. Thanks to all of you for your contributions to Rosetta@home!
8 Aug 2010, 0:00:00 UTC
· Discuss
Archived news.
We'd like to thank everyone for the recent dramatic increase in throughput. Our sustained teraflops has been at an all time high and it couldn't have come at a better time with CASP9 in progress and exciting research developments in the lab.
18 May 2010, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 2.14. For details and to report bugs, go to this thread.
10 May 2010, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 2.11. For details and to report bugs, go to this thread.
26 Apr 2010, 0:00:00 UTC
· Discuss
Archived news.
We are gearing up for CASP9 (Critical Assessment in Protein Structure Prediction) and we'll be using Rosetta@home extensively. See the announcement for more details. - Mike
26 Apr 2010, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been used to design a protein which may neutralize the flu virus. See David Baker's journal for details.
19 Apr 2010, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 2.10. For details and to report bugs, go to this thread.
12 Apr 2010, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 2.05. For details and to report bugs, go to this thread.
13 Jan 2010, 0:00:00 UTC
· Discuss
Archived news.
The project was shut down temporarily to make modifications to the SAN filesystem. -KEL/DOVA
23 Dec 2009, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 2.03. For details and to report bugs, go to this thread.
14 Dec 2009, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 2.02. For details and to report bugs, go to this thread.
5 Nov 2009, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 2.00. For details and to report bugs, go to this thread.
5 Nov 2009, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 1.98. For details and to report bugs, go to this thread.
14 Oct 2009, 0:00:00 UTC
· Discuss
Archived news.
Our filesystem became bogged down late last night. Thanks to Keith, our systems administrator, the project is back online.
16 Sep 2009, 0:00:00 UTC
· Discuss
Archived news.
Based on the current rate of data crunching, the server lag problem should be alleviated through this weekend.
11 Sep 2009, 0:00:00 UTC
· Discuss
Archived news.
The validator and scheduler servers are currently slowly processing a large work unit. We have reprioritized the WU after finding that it is causing server problem. However, it will take a while for the existing jobs to clean out. Meanwhile, server lags are expected.
10 Sep 2009, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 1.96 to solve the packaging issue in 1.95. For details and to report bugs, go to this thread.
21 Aug 2009, 0:00:00 UTC
· Discuss
Archived news.
The project is offline for the moment as we deal with an error in the recent application upate. Hopefully we will have the project back online within the next hour or so. Sorry for any inconvenience.
21 Aug 2009, 0:00:00 UTC
· Discuss
Archived news.
A number of users are still getting signature verification errors with the database file so we've updated the minirosetta application to version 1.91 with a new database filename. 1.91 is a copy of 1.90 with the only difference being a new name for the database file to ensure that every host updates the signature. To report bugs, go to this thread.
6 Aug 2009, 0:00:00 UTC
· Discuss
Archived news.
We'd like to welcome new users from the recently released Progress Thru Processors Facebook application. We'll be posting more information about current research so please stay tuned.
3 Aug 2009, 0:00:00 UTC
· Discuss
Archived news.
The project will be offline for a bit as we work on adding a couple web servers to the mix to help reduce the load on our servers.
30 Jul 2009, 0:00:00 UTC
· Discuss
Archived news.
A few details on what was done to fix the problems in the last few days are posted here.
30 Jul 2009, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 1.90. For details and to report bugs, go to this thread.
30 Jul 2009, 0:00:00 UTC
· Discuss
Archived news.
We have identify the bug that causes the program to slow down. We're currently working on updating the client. This should also solved the instant quitting problem in the last 1-2 days.
30 Jul 2009, 0:00:00 UTC
· Discuss
Archived news.
Due to a massive slowing down of the lastest code, we reverted the application back to a version equivalent to 1.82, while we work on the code. Sorry about the outage.
29 Jul 2009, 0:00:00 UTC
· Discuss
Archived news.
We've had a problem with the signatures on the 1.86 update. This issue should hopefully be resolved now with 1.87. Sorry about the outage.
16 Jul 2009, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 1.86. For details and to report bugs, go to this thread.
16 Jul 2009, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 1.82. For details and to report bugs, go to this thread.
14 Jul 2009, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 1.80. For details and to report bugs, go to this thread.
22 Jun 2009, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 1.76. For details and to report bugs, go to this thread.
16 Jun 2009, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 1.75. For details and to report bugs, go to this thread.
12 Jun 2009, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 1.71. For details and to report bugs, go to this thread.
26 May 2009, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: the project will be down for maintenance starting at around 11:00AM PST tomorrow the 20th. It may take a few hours. We will be replacing some memory for our database server and doing a full backup of the database.
19 May 2009, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 1.64. For details and to report bugs, go to this thread.
29 Apr 2009, 0:00:00 UTC
· Discuss
Archived news.
We had a brief hiccup today with our filesystem which brought the project down for a few hours. The redundancy in the Polyserve file server system that handles our BOINC data failed during a routine maintenance window. No outage was expected, and it took a long time to get the multiple servers communicating again.
28 Apr 2009, 0:00:00 UTC
· Discuss
Archived news.
Our project received a large donation from an anonymous donor and following the donor's suggestion, the University of Washington created a special Rosetta@home fund which makes it easy to make a tax deductible donation directly to our project. We've added a link to more information on our homepage in the "Join Rosetta@home" navigation section. Also, for more information see David Baker's recent journal post.
23 Mar 2009, 0:00:00 UTC
· Discuss
Archived news.
Take a look at the recent work you've made possible with your contributions! D. Baker discusses the six (6!) manuscripts that the group is publishing, having used the computing power-house that is R@H, in the Rosetta@home Journal.
11 Mar 2009, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: the project will be down again for maintenance starting at 2:00 PST today. It may take a few hours.
2 Mar 2009, 0:00:00 UTC
· Discuss
Archived news.
We're back online with a brand new database server !
26 Feb 2009, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: the project will be down for maintenance starting at 2:00 PST today. We will be upgrading the database server which may take a few hours.
25 Feb 2009, 0:00:00 UTC
· Discuss
Archived news.
Our server's back up, minirosetta 1.54 is proving to be rather stable and we got 2.5M workunits lined up - come and get them while they're fresh!
18 Feb 2009, 0:00:00 UTC
· Discuss
Archived news.
A fileserver/SAN hiccup caused the entire project to pause for a number of hours. Sorry for the outage. -KEL
16 Feb 2009, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 1.54. For details and to report bugs, go to this thread. This update is a major bugfix update addressing many issues brought forward by users over the last months. Your feedback is highly appreciated!
26 Jan 2009, 0:00:00 UTC
· Discuss
Archived news.
Well today has not been a good day for R@h. The external interface for our download/upload servers and the Ralph@home project is currently down. We will not be able to fix this until early tomorrow. Sorry for any inconvenience. Looks like it's going to be a busy day for our servers tomorrow.
6 Jan 2009, 0:00:00 UTC
· Discuss
Archived news.
Our database server lost power accidentally as work was being done on the rack. We are back up and running now.
6 Jan 2009, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 1.47. For details and to report bugs, go to this thread. Happy Xmas !
15 Dec 2008, 0:00:00 UTC
· Discuss
Archived news.
We are now recommending systems with at least 512MB of memory. The majority of tasks will run fine with 256MB but some tasks will involve larger proteins that will use more memory.
10 Dec 2008, 0:00:00 UTC
· Discuss
Archived news.
Due to a limitation on command line length set by BOINC, the jobs with name *_ZN_ABRELAX_* have their command lines automatically truncated when sent out on Rosetta@Home.This caused all the WUs to fail right away. We have canceled all these WUs and are currently investigating why they were not caught on our testing server in the first place. Sorry about any incovenience and this is a new lesson for us to learn. For details, please see this thread.
8 Dec 2008, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 1.45. For details and to report bugs, go to this thread.
4 Dec 2008, 0:00:00 UTC
· Discuss
Archived news.
The boss, David Baker, won the Raymond & Beverly Sackler International Prize in Biophysics, thanks in no small part to your contribution via Rosetta@home. Read more at http://uwnews.org/uweek/article.aspx?id=45635.
4 Dec 2008, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home is today's featured article on Wikipedia. We'd like to thank all the contributors that made this possible and those that have made the Wikipedia article such a wonderful resource.
We'd also like to welcome all the new users that have discovered Rosetta@home through the featured article.
1 Dec 2008, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: the project will be down for maintenance starting at 2:00 PST today.
On an unrelated note: WE ARE PLANNING TO INCREASE THE DEFAULT RUN TIME TO 6 HOURS AND THE MINIMUM TO 3 TO REDUCE THE LOAD ON OUR SERVERS. When and how this will occur has not yet been decided. If you have any concerns or would like to add to the discussion please post to this thread.
14 Nov 2008, 0:00:00 UTC
· Discuss
Archived news.
Our fileserver crashed last night. We are planning to upgrade the system very soon. Sorry for any inconvenience.
14 Nov 2008, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 1.40. This version contains improved protein-protein docking along with small-molecule ligands and the capabilities to do design, where the surface amino-acid residues are allowed to mutate during the simulation. Messages on the scientific aspects will be posted on the message board. Please report problems in this thread
6 Nov 2008, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 1.39. In this version, we added two important applications to minirosetta, docking and protein folding with explicit zinc metal ion. New features have been added to the graphic to show chain colors for a multi-chain protein complex and display metal ions by sphere. Please report problems in this thread. Thanks.
 |
 |
28 Oct 2008, 0:00:00 UTC · Discuss
Archived news.
We're having increasing problems with our project's fileserver, causing intermittent outages. We're in the process of building out the replacement system but it is going to take some time to iron out the issues. We apologize for the troubles.
18 Oct 2008, 0:00:00 UTC
· Discuss
Archived news.
We are currently participating to the first competition for modeling the structure of an important biological receptor. This protein is much larger than all the targets submitted so far and the calculation should last around 3 hours per model on a normal machine. The jobs start with 'AA2A.' If you have a short run time preference and you notice these jobs running past it, please do not abort them.
14 Sep 2008, 0:00:00 UTC
· Discuss
Archived news.
The Italian translation of this site is now available thanks to Manuel L, Drake VC, boboviz, Venturini D, and Ducati 749 of the BOINC.Italy team.
8 Sep 2008, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 1.32. Release information is available in this post. Please report problems in this thread.
The recent down time was due to our fileservers crashing. We are still looking into the cause.
5 Aug 2008, 0:00:00 UTC
· Discuss
Archived news.
The rosetta application has been updated to version 5.98. This version includes a bug fix for the stalled client issue. Please report problems in this thread.
25 Jun 2008, 0:00:00 UTC
· Discuss
Archived news.
As many of you have likely noticed, our throughput has decreased significantly in the last week. This was due to a couple bad batches that were sent out last week for CASP8 that were not caught by our Ralph tests. The issue was rather insidious in that the tasks would cause some clients to stall and sit idle so we were not immediately aware of the problem. The task names started with "t405". If you have any of these tasks on your client, PLEASE ABORT THEM. These batches have been cancelled. We are currently testing a fix on Ralph and an update will soon be posted. Sorry for any inconvenience.
24 Jun 2008, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 1.28. This version contains a fix for the graphics application and jumping and loop modeling protocols. Please report bugs in this thread.
10 Jun 2008, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 1.25. This version contains a fix for a file formatting problem that was causing certain jobs running abinitio folding to fail. Please report bugs in this thread.
24 May 2008, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 1.24. This version contains a fix for a memory leak that was causing tasks with long run times to fail. Please report bugs in this thread.
21 May 2008, 0:00:00 UTC
· Discuss
Archived news.
CASP8 is ramping up with a handful of targets released last week, and many more expected this week. The reason you haven't seen many CASP8 work units as we are making a final benchmarking of all the new structure prediction protocols we have developed over the past two years in the new miniRosetta code framework. The testing is on CASP7 targets--we want to determine which of our new methods are best before going all out on the CASP8 targets (yes, we would like to have completed the benchmarking before casp8 started, but there has been so much to do!). the testing should be complete monday and tuesday, and based on these results we will be sending out primarly casp8 work units next week using the approaches the tests currently running show are most successful.
I can't emphasize enough how critical all of your contributions are to our efforts to improve protein structure prediction methods! we have had many ideas on potential improvements which we have implemented, but the only way to know whether we are actually getting closer to the truth is to do rigorous testing (the casp7 targets for which we now know the structures are a perfect test set), and this would quite simply be impossible without your contributions! -- David Baker
from DB's Journal
14 May 2008, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 1.19. This version introduces several bugfixes for our CASP8 abinitio and template-based modeling protocols. We'll be testing small batches of workunits this evening and tomorrow morning. For details, see this thread, and please report bugs in this thread.
5 May 2008, 0:00:00 UTC
· Discuss
Archived news.
The minirosetta application has been updated to version 1.15. This version introduces an improved method for modeling larger RNA molecules. For details, see this thread, and please report bugs in this thread.
23 Apr 2008, 0:00:00 UTC
· Discuss
Archived news.
Rosetta has been updated to version 5.96. This version introduces an improved method for modeling larger RNA molecules. For details, see this thread.
13 Mar 2008, 0:00:00 UTC
· Discuss
Archived news.
Rosetta has been updated to version 5.95. This version includes a new method for searching beta sheet topologies. For details, see this thread.
13 Mar 2008, 0:00:00 UTC
· Discuss
Archived news.
Minirosetta has been updated to version 1.09. New graphics for windows have been included, and graphics for the mac should be in by the next version. This update also includes a variety of new experimental protocols for fullatom minimization. Please post any issues/bugs in this thread.
12 Mar 2008, 0:00:00 UTC
· Discuss
Archived news.
A new application called 'minirosetta' has been released. This application is a complete restructuring of the current rosetta applicaton and was designed to facilitate future development and science. Our goal is to gradually transition to this application as it matures. Graphics are not yet available but are currently under development. Please post any issues/bugs in this thread. We'll be issuing tasks in relatively small batches soon.
5 Feb 2008, 0:00:00 UTC
· Discuss
Archived news.
Rosetta has been updated to version 5.93. This version includes improved treatment of dihedral symmetry in rosetta. For details, see this thread,
4 Jan 2008, 0:00:00 UTC
· Discuss
Archived news.
The linux executable for Rosetta has been updated to version 5.91, to fix an issue with the workunit timing. Thanks to lots of linux users for posting and to helping us fix this quickly! For details, see this thread,
21 Dec 2007, 0:00:00 UTC
· Discuss
Archived news.
Rosetta has been updated to version 5.90. Changes include a reduction in virtual memory usage and a better exploration of proteins with complex symmetries. For details, see this thread,
20 Dec 2007, 0:00:00 UTC
· Discuss
Archived news.
Rosetta has been updated to version 5.89. Changes allow us to model proteins with complex symmetries, and improvements in the energy function used to simulate RNA. Please post any issues with the server and site in this thread.
13 Dec 2007, 0:00:00 UTC
· Discuss
Archived news.
We're up and running again. A complete update of the scheduler and web site was made so please post any issues with the server and site in this thread.
3 Dec 2007, 0:00:00 UTC
· Discuss
Archived news.
It looks like we've a left-over problem from the other day - we're working on this now.... .
3 Dec 2007, 0:00:00 UTC
· Discuss
Archived news.
The Linux executable has been updated for Rosetta@home; this fixes a small error in the prior update. For details, see this thread.
28 Nov 2007, 0:00:00 UTC
· Discuss
Archived news.
Our web site has been updated with the latest BOINC version. Please post any bugs or issues in this thread.
21 Nov 2007, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.85. This version contains some fixes in the modeling of symmetric complexes and a much improved RNA energy function. For details, see this thread.
20 Nov 2007, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down for maintenance tomorrow, Nov 21st, from around 12-2pm PST.
20 Nov 2007, 0:00:00 UTC
· Discuss
Archived news.
Outage Update: The various servers have been updated.....
15 Nov 2007, 0:00:00 UTC
· Discuss
Archived news.
We are back online. Unfortunately, we ran into some issues trying to upgrade the server so the upgrade will be postponed until early next week.
9 Nov 2007, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down tomorrow for a few hours starting at around noon Friday Nov 9th for maintenance and a server upgrade.
8 Nov 2007, 0:00:00 UTC
· Discuss
Archived news.
Due to some bookeeping issues, the current stable version of Rosetta 5.69 is now 5.82.
5 Nov 2007, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.81. This version contains small, but essential changes to the scientific protocols. For details, see this thread.
5 Nov 2007, 0:00:00 UTC
· Discuss
Archived news.
An article about Rosetta@home is in Nature. Congrats and thank you to all the volunteers that made this possible!
17 Oct 2007, 0:00:00 UTC
· Discuss
Archived news.
The project is back online.
18 Sep 2007, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down today from around 2pm to 5pm PST for maintenance.
18 Sep 2007, 0:00:00 UTC
· Discuss
Archived news.
Update: The RAID is rebuilt and now we're chasing down the next bottleneck....
18 Sep 2007, 0:00:00 UTC
· Discuss
Archived news.
Problems solved! Rosetta@home has been updated to version 5.80. This version contains support for having small molecules present in protein-protein docking simulations. This is used in the current round of Capri. For details, see this thread.
13 Sep 2007, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home is currently down due to a technical problem. We are working on a solution.
13 Sep 2007, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.78. This version includes some small but useful updates for RNA folding simulations and some special symmetric folding runs as well. For details, see this thread.
2 Sep 2007, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.77. The stable release has also been updated to 5.69. Both versions include a bug fix involving the scoring logic for closing chainbreaks. Please report any bugs in this thread.
21 Aug 2007, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.76. This version includes updates for the RNA high resolution mode and for protein workunits that involve enforcing strand pairs and then closing accompanying chain breaks ('jumping'). For details, see this thread.
12 Aug 2007, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.73. This version fixes an issue in high resolution modeling of RNA. For details, see this thread.
31 Jul 2007, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.72. This version includes some new features that will let us test variations of the energy function for proteins and RNA. For details, see this thread.
23 Jul 2007, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down tomorrow, Friday the 29th, from around 3pm to 5pm PST for maintenance.
28 Jun 2007, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.70. This version includes some new features in the RNA and symmetry modes. We are also testing the possibility of running some workunits with the old app (5.68) and another set of new workunits with the new app (5.70) to help us continue long-term studies and test new features simultaneously. For details, see this thread.
26 Jun 2007, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.68. This version allows us to carry out all-atom refinement for really large symmetric proteins where we simulate only subsystems, fixes a small bug in the RNA mode, and will allow a new sort of RNA simulation with lots of imposed base pairs.
1 Jun 2007, 0:00:00 UTC
· Discuss
Archived news.
A Japanese translation of Rosetta@home is now available. We'd like to thank Shinji Yamamoto for the translation.
1 Jun 2007, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.67. This version includes some extra features that will give us better control over the energy function for RNA workunits. For details, see this thread.
24 May 2007, 0:00:00 UTC
· Discuss
Archived news.
We forgot to mention that version 5.64 also includes 64bit applications for windows and linux platforms. These rosetta applications are copies of the 32bit version.
7 May 2007, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.64. This version includes checkpointing for pose 'fold and dock' jobs and fixes a problem that affected old PowerPC Macs. For details, see this thread.
6 May 2007, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.62. This version includes checkpointing for pose and jumping jobs and a fix for the percent complete display. For details, see this thread.
2 May 2007, 0:00:00 UTC
· Discuss
Archived news.
The project is back online.
13 Apr 2007, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down today for 2-3 hours at around 3pm PST for maintenance.
13 Apr 2007, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home is now at version 5.59! This version restores Rosetta for some Macs that were having trouble, has a more accurate estimation of ''percentage completed'' for each workunit, and has a new science mode for ''Folding and Docking'' . For more details, see this post.
2 Apr 2007, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home is now at version 5.54. This version fixes the RNA mode to follow your CPU run time preferences, rather than always producing 30 structures. There are also some small fixes in the graphics. For more details, see this post.
20 Mar 2007, 0:00:00 UTC
· Discuss
Archived news.
The project is back online.
16 Mar 2007, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down today for 2 hours at around 3pm PST for maintenance.
16 Mar 2007, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.51 -- our first update after CAPRI. This version has a nifty new 'cartoon' look for graphics. We'll also be testing a new mode to predict structures of RNA, an older cousin of DNA, that folds into interesting unique structures. For more details, see this post.
 |
 |
12 Mar 2007, 0:00:00 UTC · Discuss
Archived news.
A promotional video about Rosetta@home has been posted on YouTube.
9 Mar 2007, 0:00:00 UTC
· Discuss
Archived news.
See David Baker's recent journal post describing plans to use Rosetta@home to computationally design enzymes for simplified biochemical cycles that sequester carbon from CO2.
2 Mar 2007, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.48. We are including a special symmetric docking protocol and a fix to the problem of frequent watchdog terminations of docking workunits. For details, see this thread.
1 Mar 2007, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.46. The new version has fixed a bug which has caused a high rate of watchdog terminations for Rosetta workunits. For details, see this thread.
12 Feb 2007, 0:00:00 UTC
· Discuss
Archived news.
We will be updating the Rosetta@home application 5.46 at around 6pm PST today. As with the last update, the application is available for download in advance. See this thread for more information.
12 Feb 2007, 0:00:00 UTC
· Discuss
Archived news.
Server update: The server and php pages have been updated. Please report any issues and bugs in this thread.
7 Feb 2007, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down intermittently today as we update the server to support new BOINC 5.8.8 client features.
6 Feb 2007, 0:00:00 UTC
· Discuss
Archived news.
Outage Resolved: The project is back online.
2 Feb 2007, 0:00:00 UTC
· Discuss
Archived news.
Our server that runs the validator and a couple assimilators went down last night. We've switched to another server but it may take some time to catch up and grant credit to pending completed work units. Outage Notice: Tomorrow at around 2pm PST, Friday, the project will be down for maintenance for 2-3 hours.
1 Feb 2007, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.45. The new version has some fixes in the graphics to make it significantly more stable and, as a result, sidechains and protein rotation has been turned back on. Some science related updates are also included. For details, see this thread.
29 Jan 2007, 0:00:00 UTC
· Discuss
Archived news.
We will be updating the Rosetta@home application at around 6pm PST today. As with the last update, the application is available for download in advance. See this thread for more information.
29 Jan 2007, 0:00:00 UTC
· Discuss
Archived news.
French translation is now available. We'd like to thank Arnaud and Thierry for making this possible. To select a language visit our language selection page.
The project is back online.
18 Dec 2006, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down today for around 2 hours starting at 4pm PST for maintenance.
18 Dec 2006, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.43. The new version has simplified the graphics in an effort to reduce graphics-related crashes for a small fraction of your users. Display of sidechains has been turned off temporarily as well as mouse rotation of the models. You can expect these features to return soon! For details, see this thread.
13 Dec 2006, 0:00:00 UTC
· Discuss
Archived news.
We will be updating the Rosetta@home application in approximately 6 hours, 8pm PST. The new executable will automatically be downloaded to your client when you start a new workunit; you don't have to do anything! We've had requests from some system administrators to have an application available for download in advance; see this thread for more information.
13 Dec 2006, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home volunteers are highlighted in a recent Howard Hughes Medical Institute article.
12 Dec 2006, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.41. The new version fixes some previously spotted bionc-related bugs and other science-related bugs which are important for proceeding with the CAPRI 11 prediction. For details, see this thread.
30 Nov 2006, 0:00:00 UTC
· Discuss
Archived news.
Happy Holidays! CAPRI -- a blind docking prediction competition has opened its round 11. Please help us to find the corect answer! For more information, see this thread.
23 Nov 2006, 0:00:00 UTC
· Discuss
Archived news.
The project is back online.
22 Nov 2006, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down for a couple hours starting at 3:00 PM PST today for maintenance.
22 Nov 2006, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.40. The update introduces a new type of simulation -- predicting the structure of amyloid fibrils ( details).
 |
13 Nov 2006, 0:00:00 UTC · Discuss
Archived news.
Rosetta@home has been updated to version 5.36. The new version significantly accelerates some of the slow workunits we were testing on 5.34. For other improvements and new features, see this thread.
24 Oct 2006, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.34. The new version will allow us to test more accurate energy functions and to be more flexible with the bond lengths and angles in the protein; see this thread.
24 Oct 2006, 0:00:00 UTC
· Discuss
Archived news.
Work unit deadlines will be extended to 10 days for new work unit batches.
16 Oct 2006, 0:00:00 UTC
· Discuss
Archived news.
Outage Resolved: The project is back online. The power upgrades have been completed.
13 Oct 2006, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down tomorrow morning, Friday, at around 6:15 AM to 8:00 AM PST due to a scheduled power upgrade at our on-campus datacenter.
12 Oct 2006, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.32 -- our first update after CASP7. The new update has two major new features: 1. protein sidechains are shown in the graphics during Rosetta full-atom simulation stage (left) details; 2. a new type of structure prediciton task -- protein-protein docking is running on Rosetta@Home now (right) details;
 | 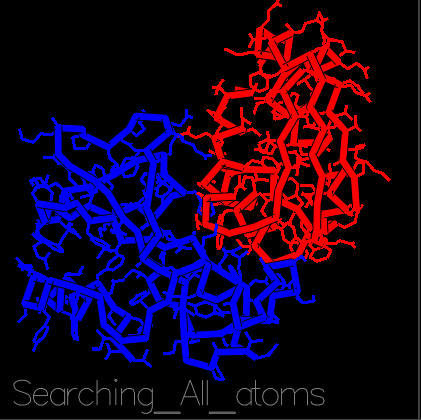 |
10 Oct 2006, 0:00:00 UTC · Discuss
Archived news.
Our servers are back online and work is flowing now.
9 Oct 2006, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down today for an hour or so at around 2pm PST for maintenance.
9 Oct 2006, 0:00:00 UTC
· Discuss
Archived news.
View results: Users can now view energy vs rmsd plots for active work units by clicking on the "Results" link under "Returning participants" and on the "Rank" numbers in the "Top participants", "Top computers", and "Top teams" pages. We'd like to thank Stuart Ozer from Microsoft Research for helping us develop this new feature. Please post comments, questions, and suggestions in this thread.
6 Oct 2006, 0:00:00 UTC
· Discuss
Archived news.
Our validator went down last night. It is now back online. There may be a delay in processing pending credits until the validator catches up with the current work flow.
24 Aug 2006, 0:00:00 UTC
· Discuss
Archived news.
Work flow is back to normal. The new work based credit values are being added to the database. They will be reported on our statistics pages tomorrow.
16 Aug 2006, 0:00:00 UTC
· Discuss
Archived news.
We are still working on the new credit system but it should be ready soon. Ralph@home is currently down for the moment as we move its hardware to another data center. It will be back online in a day or so. Thanks for your patience.
16 Aug 2006, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down for a few hours tomorrow starting at around 1pm PST to implement a new crediting system. Information about the new system is available in this Ralph@home thread.
15 Aug 2006, 0:00:00 UTC
· Discuss
Archived news.
Maintenance is complete and we are back to normal operation.
11 Aug 2006, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down today for an hour or so at around 2pm PST for maintenance.
11 Aug 2006, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home on the air!: Our local NPR station, KUOW has broadcast a story about R@H and your contributions. Listen and/or Read the story here! {Listen to DB and DK in action!!}
25 Jul 2006, 0:00:00 UTC
· Discuss
Archived news.
We are up and running now.
21 Jul 2006, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down today for an hour or so at around 3pm PST for maintenance.
21 Jul 2006, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home in the news: Rosetta@home is featured in a Seattle Times article about the Gates' Foundation awarding of $287 million for HIV vaccine research - we particularly enjoyed the phrase 'a wild-haired David Baker, a University of Washington biochemist and wunderkind'...
19 Jul 2006, 0:00:00 UTC
· Discuss
Archived news.
For anyone interested in seeing their protein structure predictions using a molecular viewing program, we've posted instructions here.
18 Jul 2006, 0:00:00 UTC
· Discuss
Archived news.
A note from David Baker: We desperately need as much CPU power as possible for the next two weeks--there are more than 25 CASP targets due, including some that are our best shots at really high resolution models. Frustratingly, we won't be able to do anywhere near as much sampling as we had planned for these proteins as there are so many coming due near the same time, and thus can't really expect the accuracy we had hoped for. So if it is at all possible for you to increase your rosetta@home cpu time for the next two weeks please do--it will make a huge difference for our collective efforts!
15 Jul 2006, 0:00:00 UTC
· Discuss
Archived news.
An article by David Baker, Proteins by Design, makes the cover story in this month's issue of The Scientist
6 Jul 2006, 0:00:00 UTC
· Discuss
Archived news.
Work is flowing again.
30 Jun 2006, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down for a couple hours today at around 4pm PST for maintenance.
30 Jun 2006, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.25. The new version has a fix for a rare checkpointing error; see this thread.
20 Jun 2006, 0:00:00 UTC
· Discuss
Archived news.
Our recommended memory requirement has been reduced to 256MB due to recent improvements in the memory footprint of our application.
21 Jun 2006, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to version 5.24. For more information on new features see this thread.
20 Jun 2006, 0:00:00 UTC
· Discuss
Archived news.
We updated our database and boinc servers today. Please report problems in this thread. The project is back online.
9 Jun 2006, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down for a few hours today starting at around 2pm PST for maintenance.
9 Jun 2006, 0:00:00 UTC
· Discuss
Archived news.
We have updated rosetta apps to version 5.22. This version has some new debugging features as well as the support for Intel Mac platform. Please read details here.
8 Jun 2006, 0:00:00 UTC
· Discuss
Archived news.
Welcome to all of our new participants! I have only very sporadic internet access as I'm out of town this next week, but I look forward to interacting with all of you here when I return. I was absolutely delighted to see the large increases of the last few days; they will really help accomplish the goals of the project! Thanks again to all of you, David.
7 Jun 2006, 0:00:00 UTC
· Discuss
Archived news.
The validator is cranking away now.
3 Jun 2006, 0:00:00 UTC
· Discuss
Archived news.
We are having problems with our validator. It should be back online later today.
3 Jun 2006, 0:00:00 UTC
· Discuss
Archived news.
We have made our first official submissions to CASP. Thanks to all who helped us predict structures for T0284 and T0287!
1 Jun 2006, 0:00:00 UTC
· Discuss
Archived news.
We have changed the workunit buffer size from 65,000 workunits to 20,000 workunits. This is so that CASP7 targets experience a smaller lag time between being queued and being sent out.
26 May 2006, 0:00:00 UTC
· Discuss
Archived news.
Work is flowing again.
24 May 2006, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down for an hour or so starting today at 3pm PST for maintenance.
24 May 2006, 0:00:00 UTC
· Discuss
Archived news.
The assimilator is back on-line.
23 May 2006, 0:00:00 UTC
· Discuss
Archived news.
The rosetta@home assimilator will be turned off from 2:30pm to approximately 4pm for fileserver maintenance. Credit for results returned in that period will be granted but may be slightly delayed.
23 May 2006, 0:00:00 UTC
· Discuss
Archived news.
We have shortened the deadline for workunits from two weeks to one week, to keep up with the tight CASP7 schedule. We will increase the deadline back to two weeks at the end of the summer, after CASP7.
22 May 2006, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home has been updated to 5.16. In addition to matching the date, the new app has slightly lower memory requirements and includes some improvements that will allow us to test different science protocols. More info here.
16 May 2006, 0:00:00 UTC
· Discuss
Archived news.
The first workunits from the CASP7 blind trials have been queued to Rosetta@home.
11 May 2006, 0:00:00 UTC
· Discuss
Archived news.
Rosetta@home updated to 5.13. New version fixes some debugging code that was causing slowdowns in a small fraction of clients.
11 May 2006, 0:00:00 UTC
· Discuss
Archived news.
Application upgraded to 5.12. New version includes the ability to send out workunit descriptions and rescaled graphics for big proteins. For more information see this thread.
9 May 2006, 0:00:00 UTC
· Discuss
Archived news.
Today's protein: T0198, a huge protein from the previous blind trial experiment CASP6.
- (Second) Lowest Energy Structure predicted by: vavega (Team XPC)
- Lowest RMSD Structure predicted by: II G II
Congratulations to today's winners!
8 May 2006, 0:00:00 UTC
· Discuss
Archived news.
Today's protein: 1b72, homeobox protein hox-b1/DNA ternary complex in homo sapiens
- Lowest Energy Structure predicted by: KWSN - Promethea (the Wanderer) (Team The Knights Who Say Ni!)
- Lowest RMSD Structure predicted by: TeAm Enterprise (Team TeAm AnandTech)
Congratulations to today's winners!
5 May 2006, 0:00:00 UTC
· Discuss
Archived news.
Today's protein: 1tul, telokin-like protein from nuclear polyhedrosis virus
- Lowest Energy Structure predicted by: NIN
- Lowest RMSD Structure predicted by: TeAm Enterprise (Team TeAm AnandTech)
Congratulations to today's winners!
30 Apr 2006, 0:00:00 UTC
· Discuss
Archived news.
Application upgraded to 5.06. New version includes a watchdog timer, and multiple checkpoints as well as some new science modes. For more information see this thread.
27 Apr 2006, 0:00:00 UTC
· Discuss
Archived news.
David Baker has been elected to the National Academy of Sciences! Congratulations, David! See this link.
25 Apr 2006, 0:00:00 UTC
· Discuss
Archived news.
Today's protein: 1bm8, DNA-binding domain of mbp1 from Saccharomyces cerevisiae (Baker's yeast)
- Lowest Energy Structure predicted by: PY-222 (Team Free-DC)
- Lowest RMSD Structure predicted by:ateale
Congratulations to today's winners!
24 Apr 2006, 0:00:00 UTC
· Discuss
Archived news.
Application upgraded to 5.01. New version includes fixes that will allow more aggressive searching, and a mechanism to abort big jobs that get restarted more than 5 times on a client.
20 Apr 2006, 0:00:00 UTC
· Discuss
Archived news.
Maintenance is complete.
18 Apr 2006, 0:00:00 UTC
· Discuss
Archived news.
Today's protein: 1fna, fibronectin, homolog from Gallus gallus (chicken)
Congratulations to today's winners!
15 Apr 2006, 0:00:00 UTC
· Discuss
Archived news.
University of Washington highlights Rosetta@home!
13 Apr 2006, 0:00:00 UTC
· Discuss
Archived news.
Today's protein: 2chf, Signal transduction protein CheY from Salmonella typhimurium
- Lowest Energy Structure predicted by: Nightlord(Team Phoenix Rising)
- Lowest RMSD Structure predicted by: necro-T-nomicon (Team FaDBeens)
Congratulations to today's winners!
12 Apr 2006, 0:00:00 UTC
· Discuss
Archived news.
Claimed credit for results with errors were granted with a maximum of 300 units.
12 Apr 2006, 0:00:00 UTC
· Discuss
Archived news.
Today's protein: 256b, cytochrome b562 (an electron transport protein) from Escherichia coli
- Lowest Energy Structure predicted by: blue_Q (Team Team MacNN)
- Lowest RMSD Structure predicted by: hidnof
Congratulations to today's winners!
8 Apr 2006, 0:00:00 UTC
· Discuss
Archived news.
The rosetta application was updated to include some new scientific code. The application version numbers have been changed to be consistent with Ralph@home. Since error rates have decreased significantly, we decided to increase the default cpu run time to 4 hours. The previous default was 2 hours.
7 Apr 2006, 0:00:00 UTC
· Discuss
Archived news.
A short story today about Rosetta@home at LiveScience.com, thanks to Benjamin Hunt (KG4ESJ).
5 Apr 2006, 0:00:00 UTC
· Discuss
Archived news.
Today's protein: 1hz6, protein L from Mus musculus (mouse)
- Lowest Energy Structure predicted by: Mephisto94 (Team L'Alliance Francophone)
- Lowest RMSD Structure predicted by: XS_DDTUNG (Team XtremeSystems)
Congratulations to today's winners!
4 Apr 2006, 0:00:00 UTC
· Discuss
Archived news.
Today's protein: 1shf, proto-oncogene tyrosine kinase, homolog from Entamoeba histolytica
- Lowest Energy Structure predicted by: Los Alcoholicos~Megaflix (Team Dutch Power Cows)
- Lowest RMSD Structure predicted by: Ziencioo (Team BOINC@Poland)
Congratulations to today's winners!
31 Mar 2006, 0:00:00 UTC
· Discuss
Archived news.
Dramatic increase in success rate: A fix for the "leave in memory bug" in the BOINC API has reduced failure rate from 9.1% to 4.2% for Windows work units with the newest Rosetta@home release.
31 Mar 2006, 0:00:00 UTC
· Discuss
Archived news.
Over 100,000 hosts have joined our project since June 2005. Thank you all for your participation! We are still actively seeking new members to help reach our target of 150 teraflops, so please spread the word out.
31 Mar 2006, 0:00:00 UTC
· Discuss
Archived news.
Today's protein: 1a19, ribonuclease inhibitor, homolog from Bacillus licheniformis
Congratulations to today's winners!
29 Mar 2006, 0:00:00 UTC
· Discuss
Archived news.
Today's protein: 1aiu, thioredoxin from humans
Congratulations to today's winners!
23 Mar 2006, 0:00:00 UTC
· Discuss
Archived news.
Today's protein: 1elw
- Lowest RMSD Structure predicted by: Markku Fager
Congratulations to today's winner! The luck of the Irish was with you!
17 Mar 2006, 0:00:00 UTC
· Discuss
Archived news.
Today's protein: 1di2
- Lowest RMSD Structure predicted by: UW-Madison CAE
Congratulations to today's winner!
15 Mar 2006, 0:00:00 UTC
· Discuss
Archived news.
Work is flowing again. We optimized database tables and backed up the database.
15 Mar 2006, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down for general maintenance starting today at 3pm PST for 1-2 hours.
15 Mar 2006, 0:00:00 UTC
· Discuss
Archived news.
We will now be posting top predictions each day!
Today's protein: 1tif
- Lowest Energy Structure predicted by: BurnHard (Team Dutch Power Cows)
- Lowest RMSD Structure predicted by: Al83
Congratulations to today's winners!
13 Mar 2006, 0:00:00 UTC
· Discuss
Archived news.
The default cpu run time is now set at 2 hours instead of 8. This change will affect new work units only.
2 Mar 2006, 0:00:00 UTC
· Discuss
Archived news.
Rosetta application update! Graphics are now available for Mac OSX platforms.
18 Feb 2006, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down starting today at 3pm PST for maintenance. The server should be back online later in the evening.
We will be updating the rosetta application today. There are a number of new features:
- Work units will have a default cpu run time of 8 hours, and users will have the option to change the cpu run time as a project specific preference. The length of work units will no longer depend on the number of predicted structures. This option was added to allow participants to reduce bandwidth usage per work unit and maintain consistent run times.
- Users will also have the option to change the frame rate and cpu use for graphics.
- A new graphics version will be available for Mac OSX users.
17 Feb 2006, 0:00:00 UTC · Discuss
Archived news.
Volunteers needed!!
We are seeking volunteers for our new alpha test project, RALPH (http://ralph.bakerlab.org).
There are a number of recent improvements to the rosetta application but we need volunteers to speed up the process of testing to make the updated application available for production as soon as possible.
If you are interested in helping to improve Rosetta@home and can spare a few extra cycles for testing, please join RALPH@home.
15 Feb 2006, 0:00:00 UTC
· Discuss
Archived news.
The validator has been fixed and work is flowing again.
30 Jan 2006, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down starting today at 8pm PST for maintenance. Our validator went down today and it remains unstable. We are going to try to fix it during the outage.
29 Jan 2006, 0:00:00 UTC
· Discuss
Archived news.
Protein Detectives, an article about David Baker and Rosetta@home, has been published recently in the HHMI Bulletin.
11 Jan 2006, 0:00:00 UTC
· Discuss
Archived news.
Work is flowing again.
6 Jan 2006, 0:00:00 UTC
· Discuss
Archived news.
We are back online.
12 Dec 2005, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down starting today at 3pm PST for maintenance. The server should be back online later in the evening.
27 Nov 2005, 0:00:00 UTC
· Discuss
Archived news.
The rosetta application versions have been updated and we are currently beta testing the screen saver version developed by Jack Schonbrun. The updated versions include new methods for increasing diversity in our search strategy and a few minor bug fixes.
11 Nov 2005, 0:00:00 UTC
· Discuss
Archived news.
We are back to normal operation. Thank you all for your patience during these recent maintenance down times.
8 Nov 2005, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down again starting at 3pm PST for around 2 hours or less to improve the performance of our database server.
8 Nov 2005, 0:00:00 UTC
· Discuss
Archived news.
Work flow is back to normal.
7 Nov 2005, 0:00:00 UTC
· Discuss
Archived news.
Outage Notice: The project will be down starting today at 3pm PST for maintenance. We should be back online later in the day.
7 Nov 2005, 0:00:00 UTC
· Discuss
Archived news.
Welcome Find-a-Drug and SETI@home users, and all new participants! We greatly appreciate your interest and participation.
A review of our research and recent progress in the field, titled Progress in Modeling of Protein Structures and Interactions, can be found in the October 28th issue of Science.
4 Nov 2005, 0:00:00 UTC
· Discuss
Archived news.
The database purging and archiving are complete and work flow is back to normal. See this post for more information.
4 Nov 2005, 0:00:00 UTC
· Discuss
Archived news.
Our database is being purged and old workunits and results are being archived. As a result, the load on the server is high and work flow has been reduced. See this thread for more information. We will be back up to speed in a day or so.
3 Nov 2005, 0:00:00 UTC
· Discuss
Archived news.
We are experiencing problems with our database and as a result the project will be down intermittently.
2 Nov 2005, 0:00:00 UTC
· Discuss
Archived news.
Rosetta designed protein, Top7, is Molecule of the Month in the Protein Data Bank.
25 Oct 2005, 0:00:00 UTC
· Discuss
Archived news.
Version 5.2.2 of the BOINC client software has been released! See the BOINC website for details.
21 Oct 2005, 0:00:00 UTC
· Discuss
Archived news.
The rosetta application has been updated on all platforms. The updated versions should have a smaller memory footprint and the OSX version should also run on 10.3.9+.
20 Oct 2005, 0:00:00 UTC
· Discuss
Archived news.
UW news - HFS helps science; you can too
14 Oct 2005, 0:00:00 UTC
· Discuss
Archived news.
We are happy to announce that this project is no longer beta. We are getting very interesting scientific results and the project is operating well thanks to user participation and input. We will post updated results soon so stay tuned!
6 Oct 2005, 0:00:00 UTC
· Discuss
Archived news.
The server and web site upgrade to BOINC 5+ is complete. Work is flowing again and the site seems okay and functional. There are some minor changes in the look and feel of the web site that were added to stay consistent with BOINC and to make future upgrades easier. Please post any significant issues that come up regarding the server and site upgrade at this forum thread.
5 Oct 2005, 0:00:00 UTC
· Discuss
Archived news.
Wednesday, October 5th, we will be upgrading our BOINC server to support 5+ clients. This site will be offline during the upgrade and will be back online the following day. You do not have to stop your client during this transition.
3 Oct 2005, 0:00:00 UTC
· Discuss
Archived news.
Welcome to the New and Returning UW Residence Hall Students!
We need your help with our research effort.
28 Sep 2005, 0:00:00 UTC
· Discuss
Archived news.
News article about our research in the Daily Telegraph.
21 Sep 2005, 0:00:00 UTC
· Discuss
Archived news.
David Baker's research in today's issue of Science. See HHMI's news release.
16 Sep 2005, 0:00:00 UTC
· Discuss
Archived news.
NIH/NIGMS article about David Baker and his research.
13 Sep 2005, 0:00:00 UTC
· Discuss
Archived news.
We have successfully switched over to a new production database server.
8 Sep 2005, 0:00:00 UTC
· Discuss
Archived news.
The server is going to be down this Wednesday through Friday for maintenance.
30 Aug 2005, 0:00:00 UTC
· Discuss
Archived news.
Over 14000 ab initio and 2800 full-atom relaxed structures made with constraints. Top full-atom relaxed model so far has an RMS of 0.54 to native. Best scoring model has an RMS of 0.70. 
18 Aug 2005, 0:00:00 UTC
· Discuss
Archived news.
New app versions added for all platforms. Old versions were removed and new ab initio and relax work units added. Folding with constraints.
23 Jul 2005, 0:00:00 UTC
· Discuss
Archived news.
Mac OS X version added.
6 Jul 2005, 0:00:00 UTC
· Discuss
Archived news.
Over 6000 structures made. So far the top model has an RMS of 2.31 to native. 
29 Jun 2005, 0:00:00 UTC
· Discuss
Archived news.
Rosetta for Windows and Linux platforms available. Initial work units are 2ptl ab initio folding tests.
26 Jun 2005, 0:00:00 UTC
· Discuss
News is available as an RSS feed

©2026 University of Washington
https://www.bakerlab.org