Posts by Admin
|
1)
Message boards :
News :
COVID-19 vaccine with IPD nanoparticles wins full approval abroad
(Message 106480)
Posted 6 Jul 2022 by Admin Post: 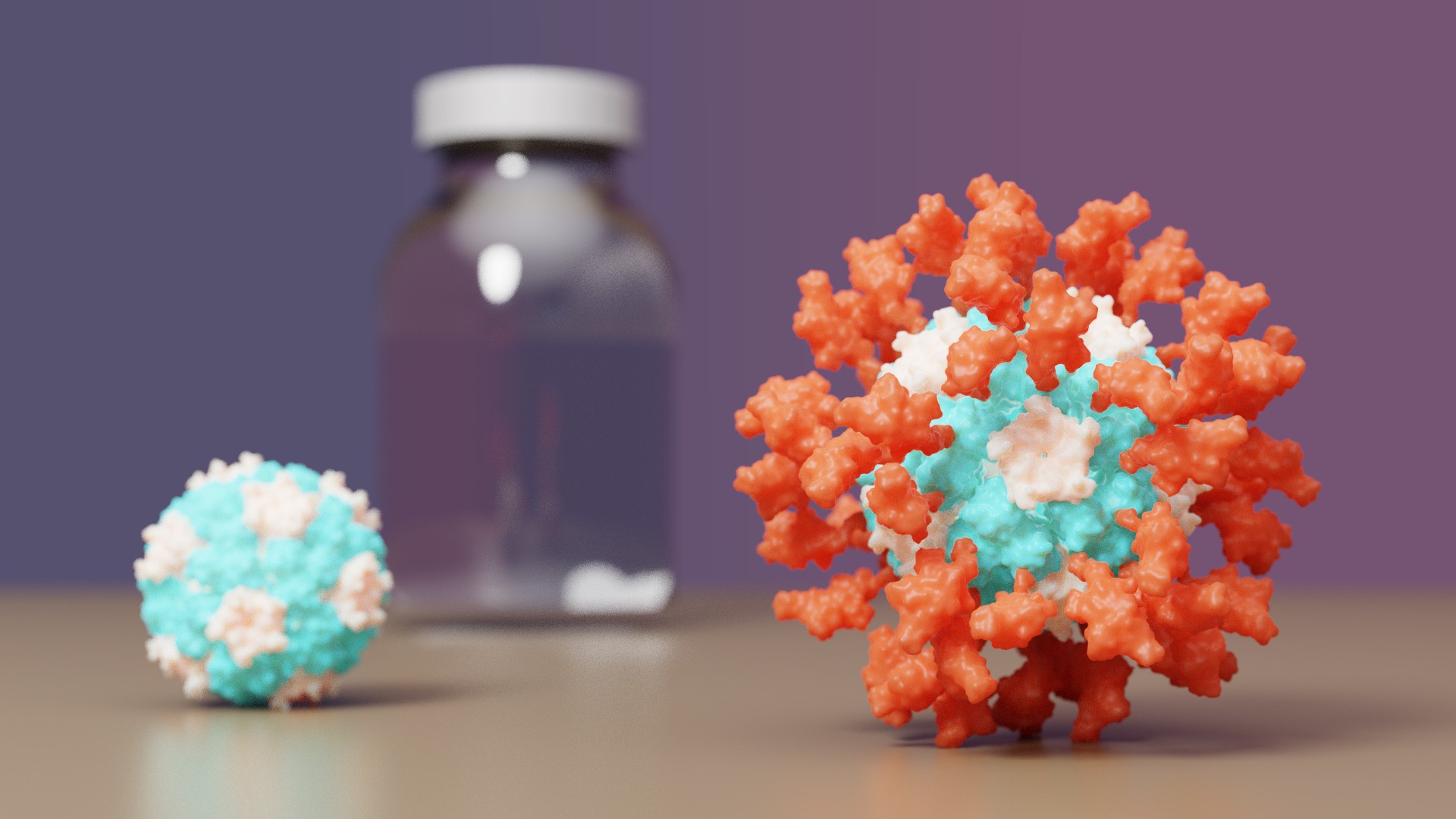 The IPD is excited to announce it's first designed protein medicine with full approval abroad. Congrats and thank you to all Rosetta@home contributors! The computing you have provided has greatly aided in de novo protein design challenges such as vaccine development leading to breakthroughs like this. For more information you can visit the IPD vaccine news post. A video is also available here. From the IPD news site: • Clinical testing found the vaccine outperforms Oxford/AstraZeneca’s • The protein-based vaccine, now called SKYCovione, does not require deep freezing • University of Washington to waive royalty fees for the duration of the pandemic • South Korea to purchase 10 million doses for domestic use A vaccine for COVID-19 developed at the University of Washington School of Medicine has been approved by the Korean Ministry of Food and Drug Safety for use in individuals 18 years of age and older. The vaccine, now known as SKYCovione, was found to be more effective than the Oxford/AstraZeneca vaccine sold under the brand names Covishield and Vaxzevria. SK bioscience, the company leading the SKYCovione’s clinical development abroad, is now seeking approval for its use in the United Kingdom and beyond. If approved by the World Health Organization, the vaccine will be made available through COVAX, an international effort to equitably distribute COVID-19 vaccines around the world. In addition, the South Korean government has agreed to purchase 10 million doses for domestic use. The Seattle scientists behind the new vaccine sought to create a ‘second-generation’ COVID-19 vaccine that is safe, effective at low doses, simple to manufacture, and stable without deep freezing. These attributes could enable vaccination at a global scale by reaching people in areas where medical, transportation, and storage resources are limited. “We know more than two billion people worldwide have not received a single dose of vaccine,” said David Veesler, associate professor of biochemistry at UW School of Medicine and co-developer of the vaccine. “If our vaccine is distributed through COVAX, it will allow it to reach people who need access.” The University of Washington is licensing the vaccine technology royalty-free for the duration of the pandemic. Congrats and thank you again to all R@h contributors! |
|
2)
Message boards :
Predictor of the day :
Predictor of the day: Congratulations to 2buiy5RiX3bN8JBr2yuiTCdV5yLJ (Team Gridcoin) for
(Message 105302)
Posted 1 Mar 2022 by Admin Post: <b>Predictor of the day</b>: Congratulations to <a href='https://boinc.bakerlab.org/rosetta/show_user.php?userid=1964246'>2buiy5RiX3bN8JBr2yuiTCdV5yLJ</a> (Team <a href='https://boinc.bakerlab.org/rosetta/team_display.php?teamid=12575'>Gridcoin</a>) for predicting the lowest energy structure for workunit <i>rb_02_26_221174_217411_ab_t000__robetta_cstwt_5.0_FT__2910548_0 <!--// -164.657 3.507 --></i> ! |
|
3)
Message boards :
Predictor of the day :
Predictor of the day: Congratulations to AKJ Surface for predicting the lowest energy structure for workunit 043_004_2_3fp9A_OB_8dd75db72f4c97fecc1c3d4a0e651a78_fold_
(Message 105211)
Posted 25 Feb 2022 by Admin Post: <b>Predictor of the day</b>: Congratulations to <a href='https://boinc.bakerlab.org/rosetta/show_user.php?userid=2083063'>AKJ Surface</a> for predicting the lowest energy structure for workunit <i>043_004_2_3fp9A_OB_8dd75db72f4c97fecc1c3d4a0e651a78_fold_SAVE_ALL_OUT_2906561_0 <!--// -146.621 7.096 --></i> ! |
|
4)
Message boards :
Predictor of the day :
Predictor of the day: Congratulations to Baxsie for predicting the lowest energy structure for workunit 174_ebeb0b76aa2934824ca392e5e407b9d0_barrel5_L1L6L8L9L4_62_fol
(Message 105196)
Posted 24 Feb 2022 by Admin Post: <b>Predictor of the day</b>: Congratulations to <a href='https://boinc.bakerlab.org/rosetta/show_user.php?userid=2125644'>Baxsie</a> for predicting the lowest energy structure for workunit <i>174_ebeb0b76aa2934824ca392e5e407b9d0_barrel5_L1L6L8L9L4_62_fold_SAVE_ALL_OUT_2906561_0 <!--// -202.612 0.977 --></i> ! |
|
5)
Message boards :
Predictor of the day :
Predictor of the day: Congratulations to Balea for predicting the lowest energy structure for workunit 145_7ed331bc664060a80a095ef321193d7a_1biaA_L5L5L4L4L6L1_55_relax
(Message 105125)
Posted 22 Feb 2022 by Admin Post: <b>Predictor of the day</b>: Congratulations to <a href='https://boinc.bakerlab.org/rosetta/show_user.php?userid=455859'>Balea</a> for predicting the lowest energy structure for workunit <i>145_7ed331bc664060a80a095ef321193d7a_1biaA_L5L5L4L4L6L1_55_relax_SAVE_ALL_OUT_2906561_0 <!--// -204.594 0.188 --></i> ! |
|
6)
Message boards :
Predictor of the day :
Predictor of the day: Congratulations to de Brus for predicting the lowest energy structure for workunit 138_6bab17d099e65dbbde1001c64660483c_3fp9A_L5L4L5L5L4L1_59_re
(Message 105101)
Posted 21 Feb 2022 by Admin Post: <b>Predictor of the day</b>: Congratulations to <a href='https://boinc.bakerlab.org/rosetta/show_user.php?userid=2098067'>de Brus</a> for predicting the lowest energy structure for workunit <i>138_6bab17d099e65dbbde1001c64660483c_3fp9A_L5L4L5L5L4L1_59_relax_SAVE_ALL_OUT_2906561_0 <!--// -224.281 0.372 --></i> ! |
|
7)
Message boards :
Predictor of the day :
Predictor of the day: Congratulations to olleoland for predicting the lowest energy structure for workunit 169_da3865ce10987d3366faf789d0cebc19_1kq1A_L1L4L4L4L3L1_63_
(Message 105053)
Posted 20 Feb 2022 by Admin Post: <b>Predictor of the day</b>: Congratulations to <a href='https://boinc.bakerlab.org/rosetta/show_user.php?userid=2137684'>olleoland</a> for predicting the lowest energy structure for workunit <i>169_da3865ce10987d3366faf789d0cebc19_1kq1A_L1L4L4L4L3L1_63_relax_SAVE_ALL_OUT_2906561_0 <!--// -243.837 0.191 --></i> ! |
|
8)
Message boards :
Predictor of the day :
Predictor of the day: Congratulations to Sanford (Team Rosetta Wisconsin) for predicting th
(Message 104997)
Posted 19 Feb 2022 by Admin Post: <b>Predictor of the day</b>: Congratulations to <a href='https://boinc.bakerlab.org/rosetta/show_user.php?userid=2139005'>Sanford</a> (Team <a href='https://boinc.bakerlab.org/rosetta/team_display.php?teamid=6463'>Rosetta Wisconsin</a>) for predicting the lowest energy structure for workunit <i>dt_denovos_post_AF25UOI_11_fragments_abinitio_SAVE_ALL_OUT_2905859_0 <!--// -124.061 2.328 --></i> ! |
|
9)
Message boards :
Predictor of the day :
Predictor of the day: Congratulations to 2buiy5RiX3bN8JBr2yuiTCdV5yLJ (Team Gridcoin) for
(Message 104932)
Posted 18 Feb 2022 by Admin Post: <b>Predictor of the day</b>: Congratulations to <a href='https://boinc.bakerlab.org/rosetta/show_user.php?userid=1964246'>2buiy5RiX3bN8JBr2yuiTCdV5yLJ</a> (Team <a href='https://boinc.bakerlab.org/rosetta/team_display.php?teamid=12575'>Gridcoin</a>) for predicting the lowest energy structure for workunit <i>dt_denovos_post_AF26MSP_05_fragments_abinitio_SAVE_ALL_OUT_2905964_0 <!--// -268.314 9.413 --></i> ! |
|
10)
Message boards :
Predictor of the day :
Predictor of the day: Congratulations to naoki (Team Governance Design Laboratory) for pred
(Message 104865)
Posted 17 Feb 2022 by Admin Post: <b>Predictor of the day</b>: Congratulations to <a href='https://boinc.bakerlab.org/rosetta/show_user.php?userid=137292'>naoki</a> (Team <a href='https://boinc.bakerlab.org/rosetta/team_display.php?teamid=19060'>Governance Design Laboratory</a>) for predicting the lowest energy structure for workunit <i>dt_native_denovo_standards7BPL_14_fragments_abinitio_SAVE_ALL_OUT_2905449_0 <!--// -310.479 4.539 --></i> ! |
|
11)
Message boards :
Predictor of the day :
Predictor of the day: Congratulations to 2buiy5RiX3bN8JBr2yuiTCdV5yLJ (Team Gridcoin) for
(Message 104838)
Posted 16 Feb 2022 by Admin Post: <b>Predictor of the day</b>: Congratulations to <a href='https://boinc.bakerlab.org/rosetta/show_user.php?userid=1964246'>2buiy5RiX3bN8JBr2yuiTCdV5yLJ</a> (Team <a href='https://boinc.bakerlab.org/rosetta/team_display.php?teamid=12575'>Gridcoin</a>) for predicting the lowest energy structure for workunit <i>dt_native_denovo_standards1xmt_fragments_abinitio_SAVE_ALL_OUT_2905272_0 <!--// -184.827 13.297 --></i> ! |
|
12)
Message boards :
Predictor of the day :
Predictor of the day: Congratulations to trail (Team ActiveFungusStudios) for predicting t
(Message 104825)
Posted 15 Feb 2022 by Admin Post: <b>Predictor of the day</b>: Congratulations to <a href='https://boinc.bakerlab.org/rosetta/show_user.php?userid=2123838'>trail</a> (Team <a href='https://boinc.bakerlab.org/rosetta/team_display.php?teamid=19739'>ActiveFungusStudios</a>) for predicting the lowest energy structure for workunit <i>dt_denovos_post_AF26LLQ_16_fragments_relax_SAVE_ALL_OUT_2905955_0 <!--// -247.795 3.792 --></i> ! |
|
13)
Message boards :
Predictor of the day :
Predictor of the day: Congratulations to 2buiy5RiX3bN8JBr2yuiTCdV5yLJ (Team Gridcoin) for
(Message 104817)
Posted 14 Feb 2022 by Admin Post: <b>Predictor of the day</b>: Congratulations to <a href='https://boinc.bakerlab.org/rosetta/show_user.php?userid=1964246'>2buiy5RiX3bN8JBr2yuiTCdV5yLJ</a> (Team <a href='https://boinc.bakerlab.org/rosetta/team_display.php?teamid=12575'>Gridcoin</a>) for predicting the lowest energy structure for workunit <i>dt_native_denovo_standards7BPL_15_fragments_relax_SAVE_ALL_OUT_2905450_0 <!--// -289.681 2.822 --></i> ! |
|
14)
Message boards :
Predictor of the day :
Predictor of the day: Congratulations to SpookyTheCat for predicting the lowest energy structure for workunit dt_native_denovo_standards2g6f_fragments_relax_SAVE_ALL_
(Message 104812)
Posted 13 Feb 2022 by Admin Post: <b>Predictor of the day</b>: Congratulations to <a href='https://boinc.bakerlab.org/rosetta/show_user.php?userid=2085916'>SpookyTheCat</a> for predicting the lowest energy structure for workunit <i>dt_native_denovo_standards2g6f_fragments_relax_SAVE_ALL_OUT_2905288_0 <!--// -164.036 0.547 --></i> ! |
|
15)
Message boards :
Predictor of the day :
Predictor of the day: Congratulations to cyleader for predicting the lowest energy structure for workunit dt_native_denovo_standards7BQE_03_fragments_relax_SAVE_ALL_O
(Message 104807)
Posted 12 Feb 2022 by Admin Post: <b>Predictor of the day</b>: Congratulations to <a href='https://boinc.bakerlab.org/rosetta/show_user.php?userid=2354309'>cyleader</a> for predicting the lowest energy structure for workunit <i>dt_native_denovo_standards7BQE_03_fragments_relax_SAVE_ALL_OUT_2905578_0 <!--// -339.828 1.297 --></i> ! |
|
16)
Message boards :
Predictor of the day :
Predictor of the day: Congratulations to University of Lynchburg Computer Science for predicting the lowest energy structure for workunit dt_native_denovo_standards6x
(Message 104797)
Posted 11 Feb 2022 by Admin Post: <b>Predictor of the day</b>: Congratulations to <a href='https://boinc.bakerlab.org/rosetta/show_user.php?userid=1044550'>University of Lynchburg Computer Science</a> for predicting the lowest energy structure for workunit <i>dt_native_denovo_standards6x1k_08_fragments_relax_SAVE_ALL_OUT_2905420_0 <!--// -246.94 2.009 --></i> ! |
|
17)
Message boards :
Predictor of the day :
Predictor of the day: Congratulations to Nekoma (Team SETI.Germany) for predicting the lowest
(Message 104773)
Posted 10 Feb 2022 by Admin Post: <b>Predictor of the day</b>: Congratulations to <a href='https://boinc.bakerlab.org/rosetta/show_user.php?userid=2241236'>Nekoma</a> (Team <a href='https://boinc.bakerlab.org/rosetta/team_display.php?teamid=26'>SETI.Germany</a>) for predicting the lowest energy structure for workunit <i>dt_native_denovo_standards1lou_fragments_relax_SAVE_ALL_OUT_2905233_0 <!--// -264.711 0.816 --></i> ! |
|
18)
Message boards :
Predictor of the day :
Predictor of the day: Congratulations to 38GfCaU9x7z7yS9E7kUmE7cyP2zA (Team Gridcoin) for
(Message 104752)
Posted 9 Feb 2022 by Admin Post: <b>Predictor of the day</b>: Congratulations to <a href='https://boinc.bakerlab.org/rosetta/show_user.php?userid=1938546'>38GfCaU9x7z7yS9E7kUmE7cyP2zA</a> (Team <a href='https://boinc.bakerlab.org/rosetta/team_display.php?teamid=12575'>Gridcoin</a>) for predicting the lowest energy structure for workunit <i>dt_native_denovo_standards1h4a_fragments_relax_SAVE_ALL_OUT_2905213_0 <!--// -254.727 0.349 --></i> ! |
|
19)
Message boards :
Predictor of the day :
Predictor of the day: Congratulations to vsral (Team Dutch Power Cows) for predicting the low
(Message 103932)
Posted 28 Dec 2021 by Admin Post: <b>Predictor of the day</b>: Congratulations to <a href='https://boinc.bakerlab.org/rosetta/show_user.php?userid=2162376'>vsral</a> (Team <a href='https://boinc.bakerlab.org/rosetta/team_display.php?teamid=78'>Dutch Power Cows</a>) for predicting the lowest energy structure for workunit <i>p53_monomer_p53_8_fragments_fold_SAVE_ALL_OUT_2855264_0 <!--// -165.342 1.317 --></i> ! |
|
20)
Message boards :
Predictor of the day :
Predictor of the day: Congratulations to Razupaltuff for predicting the lowest energy structure for workunit p53_monomer_p53_13_fragments_fold_SAVE_ALL_OUT_2855241_0
(Message 103922)
Posted 27 Dec 2021 by Admin Post: <b>Predictor of the day</b>: Congratulations to <a href='https://boinc.bakerlab.org/rosetta/show_user.php?userid=1905404'>Razupaltuff</a> for predicting the lowest energy structure for workunit <i>p53_monomer_p53_13_fragments_fold_SAVE_ALL_OUT_2855241_0 <!--// -159.601 2.628 --></i> ! |
Next 20

©2024 University of Washington
https://www.bakerlab.org