Coevolution at the proteome scale
Message boards : News : Coevolution at the proteome scale
| Author | Message |
|---|---|
|
Admin Project administrator Send message Joined: 1 Jul 05 Posts: 5146 Credit: 0 RAC: 0 |
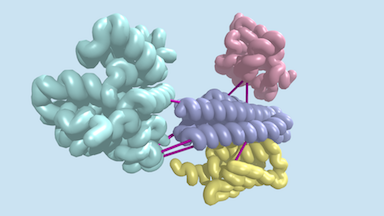 Last week, a report was published in Science describing the identification of hundreds of previously uncharacterized protein–protein interactions in E. coli and the pathogenic bacterium M. tuberculosis. These include both previously unknown protein complexes and previously uncharacterized components of known complexes. This research was led by postdoctoral fellow Qian Cong and included former Baker lab graduate student Sergey Ovchinnikov, now a John Harvard Distinguished Science Fellow at Harvard. Rosetta@home was used for much of the computing required for this work. Congratulations and thank you to all R@h volunteers. For more information about this work click here. |
![View the profile of [VENETO] boboviz Profile](https://boinc.bakerlab.org/rosetta/img/head_20.png) [VENETO] boboviz [VENETO] bobovizSend message Joined: 1 Dec 05 Posts: 2151 Credit: 12,881,353 RAC: 3,221 |
Rosetta@home was used for much of the computing required for this work. Congratulations and thank you to all R@h volunteers. Great!! |
|
trần tính Send message Joined: 29 Aug 19 Posts: 1 Credit: 0 RAC: 0 |
Great!! |
|
Bryn Mawr Send message Joined: 26 Dec 18 Posts: 440 Credit: 15,189,162 RAC: 1,294 |
Thank you, it’s wonderful to know some of the wins coming out of our efforts :-) |
|
Dad's computer Send message Joined: 22 Aug 19 Posts: 2 Credit: 935,495 RAC: 0 |
Research by Professors Charles Swanton and Sergio Quezada, University College London on project TRACERx found that cancer cells uniquely have a clonal neoantigen on their surface which normal cells do not. As we know cancer is an incredibly heterogeneous disease making it so hard to treat effectively. The findings by Swanton and Quezada would appear to have found this disease's Achilles heel an amazing discovery a long time in the waiting? I do not fully understand the science but could your work allow the manufacture of a protein capable of attacking and killing cancer cells identified by these clonal neoantigens without damaging health cells? We so desperately and urgently need new approaches to tackling this disease and I do hope your work combined with that of your international colleagues can quickly bring new drugs to the bedside and thereby help end some of the terrible suffering caused by cancer and other life threatening diseases. |
Message boards :
News :
Coevolution at the proteome scale

©2026 University of Washington
https://www.bakerlab.org